
MetaboNews
Issue 9 - April 2012
CONTENTS:
Online version of this newsletter:
http://www.metabonews.ca/Apr2012/MetaboNews_Apr2012.htm
 |
| MetaboNews Issue 9 - April 2012 |
|
CONTENTS: |
|
|
Note: Our
subscriber list is managed using Mailman, the GNU Mailing List
Manager. To subscribe or unsubscribe, please visit https://mail.cs.ualberta.ca/mailman/listinfo/MetaboNews
Current and back issues of this newsletter can be viewed from the newsletter archive (http://www.metabonews.ca/archive.html).
 |
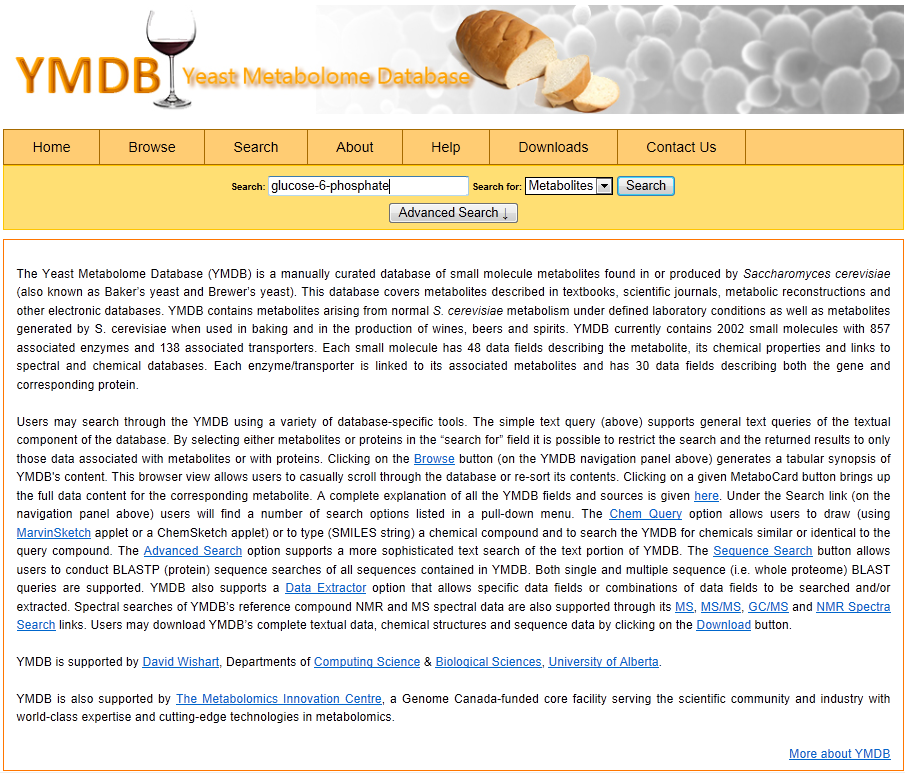
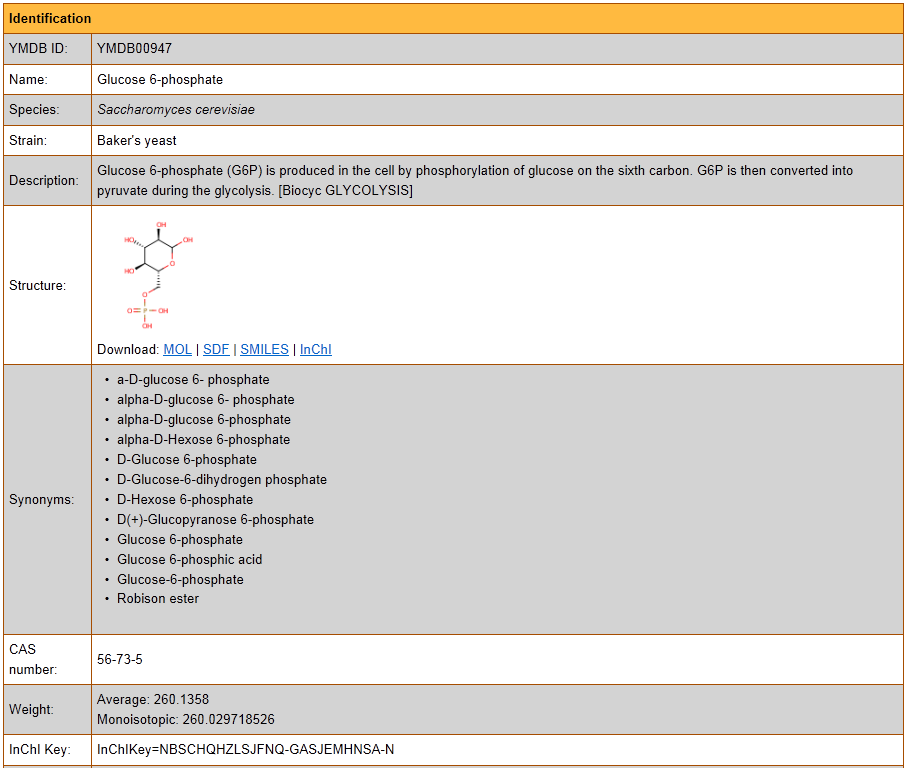
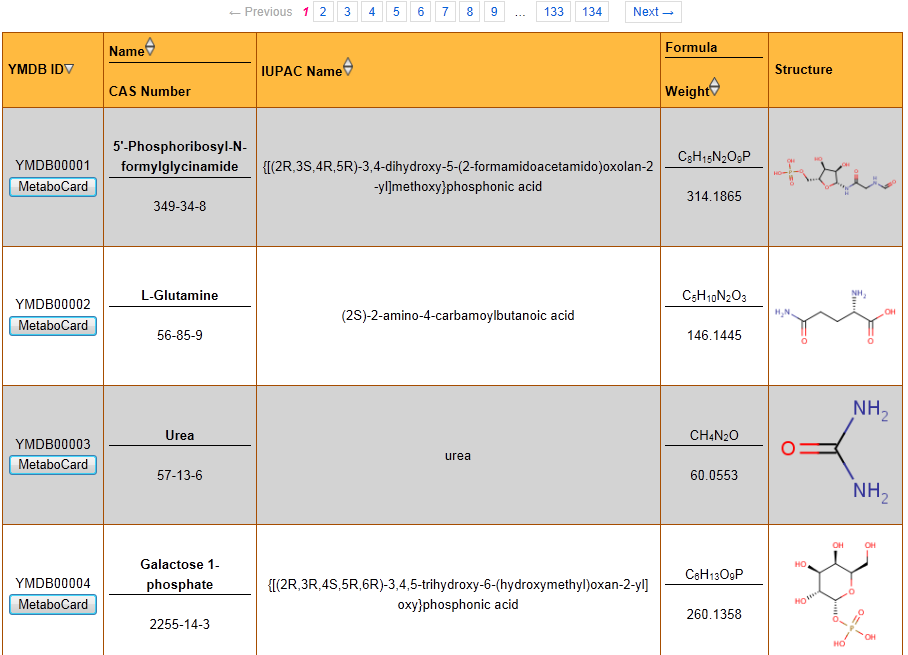
1) Software Spotlight
|
 |
| |
2) Biomarker Beacon
|
 |
3) Metabolomics Current Contents
|
 |
4) MetaboNews |
| 23 Apr 2012 |
Cancer Metabolomics:
Elucidating the Biochemical Programs that Support
Cancer Initiation and Progression [Meeting Review]
Meeting Overview In the early part of the 20th century, German biochemist Otto Warburg observed that tumor tissues and normal tissues metabolize glucose differently. Instead of relying on the citric acid cycle to extract maximal energy from a molecule of glucose, tumor cells rapidly convert glucose to lactate through glycolysis even when oxygen is abundant. Known as the Warburg effect, this observation sits at the core of the field now known as cancer metabolomics. But for many years after Warburg's observation, scientists focused on separate mechanisms of oncogenesis without considering that these metabolic differences could drive the formation of new cancers or could speed their growth. More recently, cancer researchers have recognized that these metabolic changes help cancer cells build the macromolecules that support the rapid growth and proliferation of tumors. Organized by Stephen S. Gross of Weill Cornell Medical College and Jennifer Henry of the Academy, the Cancer Metabolomics: Elucidating the Biochemical Programs that Support Cancer Initiation and Progression symposium held on February 3, 2012, is the latest in a series of meetings about cancer metabolomics. In an initial keynote address, Craig Thompson of Memorial Sloan Kettering Cancer Center gave an overview of the field and of the emerging links between oncogenic and metabolic pathways. Gary Siuzdak of Scripps Research Institute described metabolomics tools and the process of finding new cancer metabolites. As one slice of her research on sirtuin enzymes, Marcia Haigis of Harvard Medical School outlined how mitochondrial forms of these enzymes may be involved in cancer growth and proliferation. Joshua Rabinowitz of Princeton University explained fatty acid metabolism in cancer cells, and Eileen White of the Cancer Institute of New Jersey described autophagy in cancer cells. In a second keynote talk, Lewis Cantley of Beth Israel Deaconess Medical Center and Harvard Medical School discussed biochemical research to understand metabolic signals in cancer. In the final talk, Steven Lipkin of Weill Cornell Medical College described research linking the mechanisms of the primary chemotherapy in colorectal cancer, 5-fluorouracil, and a drug being tested for chemoprevention, difluoromethylornithine. Presentations available from: Lewis C. Cantley, PhD (Beth Israel Deaconess Medical Center and Harvard Medical School) Marcia C. Haigis, PhD (Harvard Medical School) Steven M. Lipkin, MD, PhD (Weill Cornell Medical College) Joshua D. Rabinowitz, PhD (Princeton University) Gary Siuzdak, PhD (The Scripps Research Institute, California) Craig Thompson, MD (Memorial Sloan-Kettering Cancer Center) |
| 23 Apr 2012 |
Warwick researchers
solve 40-year-old Fourier Transform Mass
Spectrometry phasing problem - Scientists at the
University of Warwick have developed a computation
which simultaneously doubles the resolution,
sensitivity and mass accuracy of Fourier Transform
Mass Spectrometry (FTMS) at no extra cost.
Researchers in the University’s Department of Chemistry have solved the 40-year-old phasing problem which allows plotting of spectra in absorption mode. This breakthrough can be used in all FTMS including FT-ICR, Orbitrap and FT-TOF instruments and will have applications in proteomics, petroleum analysis, metabolomics and pharmaceutical analysis among other fields. Professor Peter O’Connor, who co-developed the method, said: “We have vastly improved the quality of data available at no extra cost. "FTMS is used extensively in the fields of pharmaceuticals, healthcare, industry, natural resources and environmental management so this breakthrough represents a real step towards improving research across the board in these areas." The method is detailed in the study Absorption-Mode: The Next Generation of Fourier Transform Mass Spectra published in the journal Analytical Chemistry. It is co-authored by Professor O’Connor, Yulin Qi, Mark Barrow and Huilin Li from the University of Warwick. |
| 10 Apr 2012 |
Metabolon Launches
Strategic Outsourcing Programs Focused on Disease
Metabolism - Metabolon,
Inc., a diagnostics and services company offering the
industry’s leading biochemical profiling technology,
announces the launch of its Disease Metabolism
Strategic Outsourcing Program.
Through this program, Metabolon’s partners will collaborate toward shared objectives such as identifying novel targets, establishing high resolution phenotypic screening platforms, elucidating drug mechanism of action, discovering and validating pharmacodynamic biomarkers for translation to the clinic and for patient stratification. Partners will have access to dedicated scientific, technical and IT staff at Metabolon, along with the company’s proprietary disease ontologies, as well as supporting visualization tools for the integration of metabolomics data with existing or data derived from such disciplines as genomics and proteomics. John Ryals, Metabolon President and CEO, said, "We are excited to launch our Strategic Outsourcing Program as it represents a step change in the way the industry will think about and use metabolomics. This initiative results from years of research and development at Metabolon and has been requested by our industrial clients to fully exploit the potential of our platform. We believe that integrating metabolomic profiling deeply at the various stages of the R&D trajectory will lead to better targets, higher quality molecules, faster time to the clinic and reduced R&D risk and cost." Metabolon has completed hundreds of projects including those in oncology and metabolic disorders, and has acquired vast institutional knowledge of the metabolic pathways and targets that are involved in the onset of disease and its progression. This new program offers a turnkey metabolism center of excellence that provides pharmaceutical and biotech partners access to the most advanced metabolomics research platforms, which serve to enhance their drug discovery and development efficiency. Metabolon’s platform yields actionable insight into key disease metabolic processes, drug action, and patient response. Source: MarketWatch |
 |
5)
Metabolomics Events
|
| 3-4 May 2012 |
Canadian Bioinformatics Workshops:
Informatics and Statistics for Metabolomics This course is intended for graduate students, post-doctoral fellows, clinical fellows and investigators who are interested in learning about both bioinformatic and cheminformatic tools to analyze and interpret metabolomics data. Prerequisite: Your own laptop computer. Minimum requirements: 1024x768 screen resolution, 1.5GHz CPU, 1GB RAM, recent versions of Windows, Mac OS X or Linux (Most computers purchased in the past 3-4 years likely meet these requirements). If you do not access to a laptop, you may loan one from the CBW. Please contact course_info@bioinformatics.ca for more information. Course Objectives The workshop will cover many topics ranging from understanding metabolomics technologies, data collection and analysis, using pathway databases, performing pathway analysis, conducting univariate and multivariate statistics, working with metabolomic databases and exploring chemical databases. Participants will be given various data sets and short assignments to assist with the learning process. Register now as space is limited to 30 participants. For more information, please visit http://bioinformatics.ca/workshops/metabolomics-workshop.
Workshop Flyer (PDF): http://www.metabonews.ca/Mar2012/events/InformaticsMetabolomicsJan2012s.pdf Canadian Bioinformatics Workshop Series Flyer (PDF): http://www.metabonews.ca/Mar2012/events/CBW_FinalPoster_2012_small.pdf
|
| 7-8 May 2012 |
LIPID MAPS Annual Meeting 2012: Lipidomics
Impact on Cell Biology, Metabolomics and
Translational Medicine The meeting program tentatively features the following six sessions:
For more information, please visit http://www.lipidmaps.org/meetings/2012annual/
|
| 20-24 May 2012 |
60th ASMS Conference on Mass Spectrometry and
Allied Topics For more information, please visit http://www.asms.org/Conferences/AnnualConference/GeneralInformation/tabid/127/Default.aspx.
|
| 21-23 May 2012 |
Les 6èmes Journées
Scientifiques du Réseau Français de
Métabolomique et Fluxomique (JS 6 RFMF)
For more information, please visit https://colloque4.inra.fr/6_js_reseau_francais_metabolomique_fluxomique.
|
| 18-21 June 2012 |
Shortcourse: Metabolic Phenotyping in Disease
Diagnosis & Personalised Health Care The course will be held on the 18th-21st June, 2012.
|
| 25-28 June 2012 |
METABOLOMICS 2012: Breakthroughs in plant,
microbial and human biology, clinical and
nutritional research, and biomarker discovery The 2012 meeting promises a program full of practical workshops and parallel sessions covering the broad range of biological and technological metabolomics topics as well as providing rich opportunities for networking. Join us as we gather together to share ideas, insights, advances and obstacles in the multi-faceted world of metabolomics. For more information, please visit http://www.metabolomics2012.org.
|
| 10 July 2012 |
Merck Deep Dive: Translational Technologies
for Basic and Clinical Scientists For more information, please visit http://www.cvent.com/events/merck-deep-dive-molecular-biomarkers-protemics-genomics-metabolomics-/event-summary-be463e01b8974ad99e3d84af4be50bef.aspx.
|
| 28-31 Aug 2012 |
NuGOweek 2012 (9th Edition): “Nutrition,
lifestyle and genes in the changing environment” Venue & Accommodation The conference venue and hotel is the hotel and conference centre Hilton Helsinki Kalastajatorppa . Scientific & Conference Committee The members of the Scientific Committee are: Matti Uusitupa (chair), Jussi Pihlajamäki, Marjukka Kolehmainen, Kaisa Poutanen, Kirsti Husgafvel-Pursiainen, Jim Kaput, Martin Kussmann, Ben van Ommen, Lars Ove Dragsted, Stine Marie Ulven, Antonio Zorzano, Lorraine Brennan The members of the Conference Organising Committee are: Marjukka Kolehmainen, Matti Uusitupa, Kaisa Poutanen, Hannu Mykkänen, Jussi Pihlajamäki, Kati Hanhineva, Vanessa de Mello Laaksonen, Fré Pepping, Ingeborg van Leeuwen-Bol. Further details about the programme will be made available end of February 2012. Course on Metabolomics Also this year we will organise a PhD course prior the NuGOweek. From 20-25 August 2012 the Course on Metabolomics 2012 will be held in Kuopio, Finland. For further information please contact Ingeborg van Leeuwen-Bol or visit http://www.nugo.org/nugoweek/42113/5/0/30. |
| 25-27 Sep 2012 |
Metabomeeting 2012 Previous meetings have been held at King's College, Cambridge, Imperial College London, the Norwich Bioscience Institutes, the Ecole Normale Supérieure de Lyon and in Helsinki in collaboration with VTT Research Centre. The Metabomeeting conference focuses on providing a forum for researchers in metabolomics to meet and discuss all aspects of its research, development and application. Confirmed speakers for the meeting include: Professor Robert Hall, Plant Research International, The Netherlands, who will present the plenary lecture. Professor Rainer Breitling, University of Glasgow, UK. Professor Hannelore Daniel, Technical University of Munich, Germany. Dr Jerome Jansen, Raboud University, Nijmegen, The Netherlands. Dr Nick Lockyer, University of Manchester, UK. Professor George Nychas, Agricultural University of Athens, Greece. Professor Ian Wilson, AstraZeneca, UK. You are invited to submit abstracts for consideration for podium presentations. All abstracts need to be submitted before May 4th 2012. More details are available via http://thempf.org/mpf_cms/index.php/conferencesworkshops/forthcoming-mpf-events/78-metabomeeting-2012. |
| 7-9 Nov 2012 |
29th LC/MS Montreux Symposium The Montreux LC/MS 2012 conference: Special highlights on Metabolomics and Clinical Chemistry The field of LC/MS is continuously growing as is reflected by the participation of over 30 nationalities and by scientific contributions from a variety of research and development domains such as pharmaceutical, biotechnological, food, environmental and research on novel instrumentation and new LC/MS fields such as nanotechnology and microfluidics, UPLC, low flow rate spray techniques, proteomics, and systems biology. In collaboration with the Metabolomics Society, a special joint parallel program for this rapidly emerging field is organized addressing the technology as well as novel systems-based biology approaches in pharma, nutrition, clinical chemistry, plant sciences, and medical biology. A parallel program is organized together with various Clinical Chemistry societies focusing on current and future LC/MS options in clinical diagnosis. Accreditation by related societies for the program as well as the short course has been applied for. For more information, visit http://www.lcms-montreux.com/. Conference flyer (PDF): http://www.metabonews.ca/Mar2012/events/flyer_LC-MS _Montreux_conference_2012.pdf |
 |
6) Metabolomics Jobs |
This is a resource for advertising
positions in metabolomics. If you have a job you would like
posted in this newsletter, please email Ian Forsythe (metabolomics.innovation@gmail.com).
Job postings will be carried for a maximum of 4
issues (8 weeks) unless the position is filled prior to that
date.
Jobs
Offered
| Job Title | Employer | Location | Date Posted | Source |
|---|---|---|---|---|
| Experimental Officer in Metabolomics | University of Birmingham | Birmingham, UK | 16-Apr-2012 | Metabolomics Society Jobs |
| Experimental Officer in Bioanalytical Chemistry | University of Birmingham | Birmingham, UK | 16-Apr-2012 | Metabolomics Society Jobs |
| Lectureship in Metabolomics / Metabolic Biochemistry | University of Birmingham | Birmingham, UK | 16-Apr-2012 | Metabolomics Society Jobs |
| Postdoc position in metabolomic NMR | University of Freiburg | Freiburg, Germany | 12-Apr-2012 | University
of Freiburg |
| PhD Research Project: Metabonomic evaluation of Lactobacillus salivarius ability to protect against Brachyspira pilosicoli-induced intestinal spirochetosis | University of Reading | Reading, UK | 31-Mar-2012 | FindAPhD.com |
| Post-doctoral Position | CIC bioGUNE Centre | Bilbao, Bizkaia, SPAIN | 26-Mar-2012 | Metabolomics Society Jobs |
| PhD Studentship in Bioanalytical Chemistry | University of Birmingham | Birmingham, UK | 22-Mar-2012 | Metabolomics Society Jobs |
| PhD fellowship: "Metabolomics of Nicotiana attenuata’s ecological interactions" | Max Planck Institute for Chemical Ecology | Jena, Germany | 21-Mar-2012 | Max Planck Institute for Chemical Ecology |
|
Ian J.
Forsythe, M.Sc.
MetaboNewsEditor Department of Computing Science
University of Alberta 221 Athabasca Hall Edmonton, AB, T6G 2E8, Canada Email: metabolomics.innovation@gmail.com Website: http://www.metabonews.ca LinkedIn: http://ca.linkedin.com/in/iforsythe Twitter: http://twitter.com/MetaboNews Google+: https://plus.google.com/118323357793551595134 Facebook: http://www.facebook.com/profile.php?id=100003143138743 |
This newsletter is
produced by The Metabolomics Innovation Centre (TMIC, http://www.metabolomicscentre.ca/)
for the benefit of the worldwide metabolomics
community.
 A single source destination for fee-for-service metabolic profiling including comprehensive metabolite identification, quantification, and analysis
|