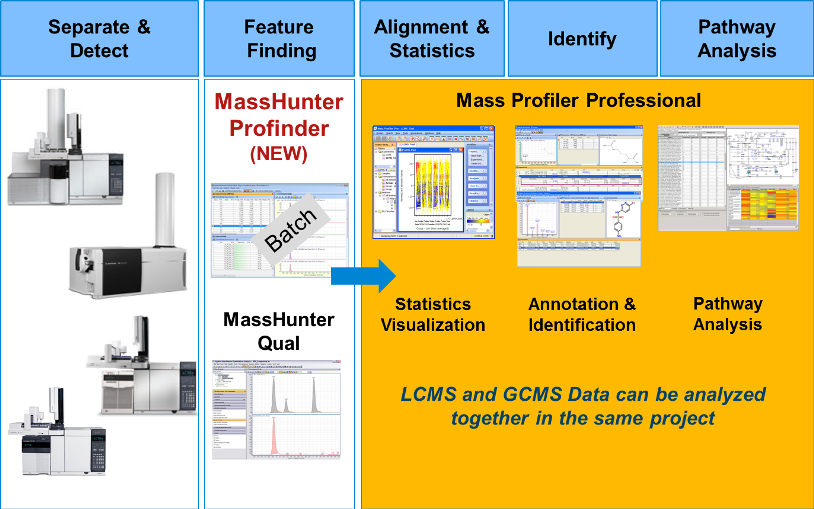
7-11 Apr 2014
|
Leiden University:
Workshop Metabolomics 2014
Basics and
Applications to Plant Sciences
Venue: Leiden, The
Netherlands
Dates
April 7 - 11,
2014, workshop
April 14 -
18, 2014, hands-on (optional)
April 21 - 25, 2014, wrapping up of
workshop and hands on (optional)
Description
and aim of the workshop
Metabolomics
has become an important tool in life
sciences, including studies of plant
interaction with its environment and
studies on the activity of medicinal
plants. The workshop is organized by the
Institute of Biology, Leiden University.
The workshop is particularly aimed at
researchers experienced with the isolation
and identification of natural products but
do not have experience yet with
metabolomics.
Topics
- Primary and
Secondary Metabolism in Plants
- Analytical Techniques: Chromatography, MS,
NMR
- Basics of
Multivariate Data Analysis
- Applications
in
Plant
Sciences
Costs
800 Euro for the
workshop.
Including handout,
lunches (April 7-11), drinks and one
workshop dinner
(hotel accommodation is
not included)
200 Euro for the
hands-on.
Including supervision
and use of the laboratory equipment
(hotel accommodation is
not included)
25 Euro for the
wrapping up.
Including supervision
(hotel accommodation is
not included)
Maximum number of participants (First
come first served)
For the workshop, April
7-11: 25 participants
For the hands-on, April
14-18: 10 participants
For wrapping up, April
21-25: 10 participants
Organized by Young Hae Choi, Jos Frantzen,
and Rob Verpoorte. For more
information, email j.frantzen@drsupport.nl.
Registration deadline: 31
March 2014
The workshop
flyer is available here.
For more details, please visit http://www.plantsandmetabolomics.nl/
|
10 Apr 2014
|
Mitochondria, Metabolism
and Disease
Venue: New York
Academy of Sciences, 7 World Trade Center,
250
Greenwich St, 40th Fl., New
York City, USA
Organizers:
Vamsi K. Mootha, MD
(Harvard Medical School), Steven
Gross, PhD
(Weill Cornell Medical College), Jennifer
Henry, PhD
(The New York Academy of Sciences)
Speakers: Robert Balaban, PhD (National
Heart, Lung, and Blood Institute, NIH), Robert
Bao, PhD (Massachusetts General Hospital), Salvatore
DiMauro, MD (Columbia University Medical
Center), Steven Gross, PhD (Weill Cornell
Medical College), Costas A. Lyssiotis, PhD
(Weill Cornell Medical College), Vamsi K.
Mootha, MD (Harvard Medical School), Jared
Rutter, PhD (University of Utah School of
Medicine)
Description: Mitochondria are the
cell's "power plants" and serve a critical role in
global metabolism. Accordingly, dysfunction or
damage of mitochondria can greatly perturb
metabolism, and underlies a diverse range of human
diseases, ranging from neurodegenerative
conditions (ALS, Alzheimer's and Parkinson's
diseases), epilepsy, psychiatric illness and
autism, to atherosclerotic heart disease, stroke,
liver disease, type 2 diabetes and cancer.
For more details, please visit www.nyas.org/MitochondrialDisease
|
28-29 Apr 2014
|
Companion Diagnostics:
From Biomarker Identification to Market Entry
Venue: New
York Academy of Sciences, 7 World
Trade Center, 250
Greenwich St, 40th Fl.,
New
York City, USA
The recent growth in complex biological data has
enabled a major paradigm shift in healthcare away
from "one-size-fits-all" approaches towards
customized, patient-tailored therapies.
Biomarker-based companion diagnostics (CDx),
designed to identify responsive patient
sub-populations or those likely to experience
adverse drug effects, lie at the heart of this
personalized, precision medicine movement.
This 2-day conference will provide a neutral forum
for international scientists and clinicians from
academia, industry, and government — as well as
healthcare policy makers, regulatory experts,
insurance sector representatives, and other
stakeholders — to discuss research, financial, and
regulatory strategies that will facilitate CDx
development and integration into clinical care.
Presentations will illustrate successes and
failures based on case studies; evaluate emerging
applications of technologies such as epigenetics,
bioinformatics, and nanotechnology; analyze
diverse therapeutic target areas including the
pioneering field of cancer along with
inflammatory, infectious, cardiovascular,
metabolic, and neurodegenerative diseases;
recognize regulatory hurdles; and formulate
solutions to better improve public health with CDx
and personalized medicine.
For more information, please visit http://www.nyas.org/CDx2014
|
30 Apr 2014
|
Analytical Tools for
Cutting-edge Metabolomics - a joint meeting of
the Analytical Division of the RSC and the
international Metabolomics Society
Venue: Chemistry Centre, Burlington House,
London, UK (Google
Map
Location)
Date: 30 April
2014, 09:30-16:45
Analytical
chemistry has been one of the driving forces
behind the development of metabolomics research
over the past decade. The conference will bring
together exceptional scientists for a program
consisting of plenary and invited talks, posters,
as well as an oral session devoted to early career
researchers. It will be an excellent opportunity
for analytical chemists to learn more about
metabolomics and its application, and for
metabolomics scientists to improve their knowledge
of cutting-edge bioanalytical tools.
Deadline
for submission of abstracts: 14
March 2014
Speaker
Information:
Prof. Jeremy Nicholson, Imperial
College, London UK - Plenary speaker
Dr Julian Griffin, MRC Human Nutrition
Research, Cambridge, UK
Prof. Roy Goodacre, University of
Manchester, UK
Prof. Jean-Luc Wolfender, University
of Geneva, Switzerland
Dr Steffen Neumann, Leibniz Institute
of Plant Biochemistry, IPB Halle,
Germany
Prof Paul Thomas, Loughborough
University
For more
details, please visit the
event
website.
|
19-21 May 2014
|
8e Journées Scientifiques
du Réseau Français de Métabolomique et
Fluxomique (RFMF)
Venue: Lyon, France
The French Metabolomics and Fluxomics Network
(RFMF) is a non-profit organization dedicated to
the development of metabolomics and fluxomics, and
the promotion of the presentation of research
achievements in these domains, in France. Since
2005, RFMF has organized a total of 7 congresses
in France (Toulouse 2005, Saint Sauves d’Auvergne
2006, Bordeaux 2008, Marseilles 2010, Paris 2011,
Nantes 2012 and Amiens 2013). The number of
laboratories and attendees has grown steadily over
the years. In October 2013, the RFMF and the
international Metabolomics Society form an
international affiliation.
The Amiens RFMF Congress (7
JS RFMF) took place in 2013 due to the
generous support of Picardie Jules Verne
University and various corporations. This event
featured three keynote speakers, Age
Smilde, Amsterdam, The Netherlands; Joachim
Kopka, Golm-Postdam, Germany and Reza Salek,
Cambridge, UK presenting state-of-the-art
topics for metabolomics. This event was very
successful, with 162 attendees from 75 public and
private laboratories, 59 high-quality scientific
presentations, 4 workshops
and 6 industrial
seminars. Four conferences, one workshop and
one industrial seminar were held in English. The
2013 event gave an opportunity to gauge the
strength and dynamic qualities of the French or
French-speaking metabolomics and fluxomics
community.
The 8th RFMF Congress will take place in Lyon
(Eastern France) in May 19-21, 2014. The
Metabolomic Community of the Lyon and
Rhône-Alpes Region laboratories is the local
organizer of the 2014 edition.
The key topics, chosen by Lyon metabolomics
community, for the 8th RFMF Congress are:
- Applications of Metabolomics and Fluxomics
in the areas of the Health (clinical
applications, epidemiology, cohort study,
neurosciences)
- Applications
of Metabolomics and Fluxomics in the
areas of Environment and Chemical
ecology
The event will include invited plenary lectures,
oral presentations, short presentations, and a
poster session. The conference languages are
French and English. Most of the abstracts and
slide presentations will be written in English.
Confirmed invited speakers include Miroslava
Cuperlovic-Culf, from Moncton, Canada, Emmanuel
Gacquerel, from Jena, Germany, Thomas
Illig, from Hannover, Germany and Mark
Viant, from Birmingham, UK.
The 8th RFMF Congress will include several
workshops, one of them dealing with the Galaxy
platform (workflow for metabolomics), one dealing
with “Sample preparation for metabolomics study”,
and a third one with “MetaboLights and COSMOS
initiative”.
RFMF is generously supporting the attendance of
students or post-docs (under 35 years old) at The
8th RFMF Congress Lyon 2014 through the provision
of travel
grants (up to €1000 for overseas
applicants).
For more information on the 8th RFMF Congress,
please visit https://colloque.inra.fr/8_js_rfmf_lyon_2014
|
22-23 May 2014
|
Metabolomics Conference -
Advances & Applications in Human Disease
Venue: Hyatt Regency Cambridge,
Cambridge, USA
The Metabolomics – Advances & Applications
in Human Disease conference will focus on
the clinical aspects in the metabolomics field
such as advances in metabolite markers for use in
personalized medicine, novel computational
approaches for metabolome assessment and discuss
developments in clinical assay
standardization/qualification for the
implementation of metabolite markers in clinical
development of drug candidates.
Special
offer for Metabolomics Society Members and
MetaboNews Subscribers:
Receive 30% off Registration with discount
Code: meta30
Distinguished Speakers
- Richard Beger, Director, Biomarkers and
Alternative Models Branch, Division of Systems
Biology, National Center for Toxicological
Research, FDA
- Susan Cheng, Associate Physician, Brigham
and Women’s Hospital, Instructor in Medicine,
Harvard Medical School
- Robert H. Weiss, Professor & Chief of
Nephrology, Sacramento VA Medical Center,
University of California, Davis
- David Wishart, Professor, Computing Science
& Biological Sciences, University of
Alberta
- View the full list
of speakers
Agenda Highlights
Keynote Presentations:
- Metabolomics of Acetaminophen Overdose in
Rodents and Children
- Quantitative Metabolomics & Medical
Biomarkers: Challenges & Opportunities
Sessions:
- Metabolite Identification – Targets and Hits
& Pathways
- Standardization & Validation
- Computational Approaches to Assessing the
Metabolome
- Advances in Metabolite Markers for
Personalized Medicine
- Technological Advances
- Clinical
Assays for Metabolite Markers
- View the detailed
agenda
This conference is co-located with the Drug
Discovery Summit 2014, which includes the
conferences:
To register, click here.
For more information, see the event
flyer or visit http://www.gtcbio.com/conference/metabolomics-overview
|
16-17 Jun 2014
|
Informatics and Statistics
for Metabolomics (2014)
Venue: Vancouver, BC, Canada
Course Objectives
A poster announcing this workshop can be found here.
The workshop will cover many topics ranging from
understanding metabolomics technologies, data
collection and analysis, using pathway databases,
performing pathway analysis, conducting univariate
and multivariate statistics, working with
metabolomic databases and exploring chemical
databases. Participants will be given various data
sets and short assignments to assist with the
learning process.
Target Audience
This course is intended for graduate students,
post-doctoral fellows, clinical fellows and
investigators who are interested in learning about
both bioinformatic and cheminformatic tools to
analyze and interpret metabolomics data.
Prerequisite: Your own laptop computer.
Minimum requirements: 1024x768 screen resolution,
1.5GHz CPU, 1GB RAM, recent versions of Windows,
Mac OS X or Linux (Most computers purchased in the
past 3-4 years likely meet these requirements). If
you do not access to a laptop, you may loan one
from the CBW. Please contact course_info@bioinformatics.ca
for more information.
Pre-Readings: You are expected to have
completed the following tutorials in R beforehand.
The tutorial should be very accessible even if you
have never used R before. Please complete the
following: R
Tutorial
Informatics and Statistics for
Metabolomics Workshop flyer: http://www.metabonews.ca/Apr2014/events/CBW/metabolomics-cbw-2014.pdf
Canadian Bioinformatics Workshop
Series flyer: http://www.metabonews.ca/Apr2014/events/CBW/flyer-cbw-2014_sm.pdf
For
more information, visit http://bioinformatics.ca/workshops/2014/informatics-and-statistics-metabolomics-2014.
|
23-26 Jun 2014
|
Metabolomics 2014: 10th
Annual International Conference of the
Metabolomics Society
The Official Joint Conference of the
Metabolomics Society and Plant Metabolomics
Platform
The Official Annual Meeting of the Metabolomics
Society
Venue: Tsuruoka, Japan
Health, medical, pharmaceutical, nutritional,
agricultural, microbial, bioenergy, environmental
and plant sciences meet biochemical, analytical
and computational technologies.
We are delighted to host the 10th Anniversary of
the International Conference of the Metabolomics
Society (Metabolomics2014) at Keio University in
Tsuruoka City, where the very first meeting of the
society was held in 2005. Since then, Tsuruoka has
grown to “a city of metabolomics”; various
additional research buildings have been built, and
two spin-out companies established. Tsuruoka is a
pretty city located 500 km north of Tokyo (about 1
hour flight), and surrounded by beautiful Japanese
nature, historic spots, and exotic culture. You
will also enjoy the best authentic Japanese food
and sake (rice wine), as well as hot springs. So,
come celebrate the 10th anniversary of the
society, and enjoy high-quality scientific
presentations by top-notch researchers around the
world.
Key Dates
- Abstract
Deadline for Oral
Presentations Extended to
midnight of April 6th 2014
(GMT)
- Early
Registration Deadline: April
10th 2014
- Deadline
for Late Poster Abstracts:
midnight of April 30th 2014
(GMT)
- Regular Registration Deadline: May
29th 2014
- Late Registration: From 1 June 2014
For
detailed information about
Metabolomics 2014, visit http://metabolomics2014.org.
|
10-12 Sep 2014
|
Metabomeeting 2014
Venue: The Royal Institution, London, UK
SELECTBIO are delighted to announce that we are
partnering with the Metabolic Profiling Forum
(MPF) to host Metabomeeting 2014. The MPF will
focus on the conference program while SELECTBIO
will take care of logistics, promotion and
exhibition/sponsorship activities.We are expecting
up to 300 attendees offering a unique opportunity
to network with key researchers who are making
innovative discoveries within this field.
Call for Papers
If you would like to be
considered for an oral
presentation at this
meeting, Submit an
abstract for review now!
Oral Presentation
Submission Deadline: 31
January 2014
Call for Posters
You can also present your
research on a poster while
attending the meeting.
Submit an abstract for
consideration now!
Poster Submission
Deadline: 27 August 2014
Agenda Topics
Applied Metabolomics
Drug Discovery and Pharma
Human Disease
Human Health and Nutrition
Microbial, Invertebrate
and Environmental
Applications
Plants
Data Analysis and
Integration with Systems
Biology
Metabolite Identification
For more
details, please visit the conference
website.
|
14-18 Sep 2014
|
International Chemometrics
Research Meeting ICRM 2014
Venue: The Golden Tulip Valmonte Hotel, Berg en
Dal near Nijmegen, The Netherlands
The ICRM conference is held once every
three/four years and is
organized by the Dutch
Chemometrics Society
(DCS), a working group of
the Royal Netherlands
Chemical Society. The aim
of this conference is to
bring together people from
a wide range of industry,
research and academic
backgrounds to share and
discuss recent
developments in the field
of chemometrics.
Chemometrics is the
discipline concerned with
the extraction of
information from
analytical chemical data.
It has numerous successful
applications in an
extremely wide range of
industries, for example in
chemical and
pharmaceutical research
and production. The
conference focuses on
presentations by prominent
speakers from around the
globe, who will be invited
to share their knowledge
both during lectures on a
wide range of topics from
the field of chemometrics,
as well as during the
extensive, succeeding
discussions. Confirmed
keynote lecturers of the
conference currently are
Alan Gelfand, John H.
Kalivas, Olav M. Kvalheim,
Iven van Mechelen, Mia
Hubert, Dennis R. Helsel
and Neil B. Gallagher.
At this ICRM 2014
conference also time has
been reserved for
contributed lectures. We
invite you to submit
abstracts for oral
contributions, as well as
for the two poster
sessions. Abstracts can be
submitted through the
conference website at www.icrm2014.org. Submission deadline
is May 1st 2014. Note that
only submissions from
registered attendees are
taken into the review
process.
Email contact: icrm2014@xs4all.nl
Registration: www.icrm2014.org/registration/registration.html.
For more
details, please visit http://www.icrm2014.org/
|
| 29-30 Oct 2014
|
Clinical Applications of
Mass Spectrometry
Venue: Barcelona, Spain
With the ability to measure multiple analytes with
high sensitivity, often faster and more cheaply
than other methods, mass spectrometry is becoming
an attractive method of analysis for the clinic.
Featuring an array of leading researchers and
clinicians, SELECTBIO’s Clinical
Applications of Mass Spectrometry conference aims
to provide you with an insight into the latest
developments in this area.
As the analytical power of mass spec is realised,
the range of applications using this technology
continues to expand. Focus at this meeting will be
given to both traditional & emerging uses of
MS in the clinic. Hot topics to be covered include
developments of MS in applications ranging from
vitamin D detection to newborn blood spot
analysis. Attending this event will provide you
with excellent opportunities for networking with
like minded peers, helping you to build new
relationships and optimise your workflow.
Running alongside the conference will be an
exhibition covering the latest technological
advances and associated services from leading
solution providers within this field. Registered
delegates will also have access to the co-located
Food Analysis Congress, ensuring a cost
effective trip.
Keynote Speakers:
- Donald
Hunt, Professor, University of Virginia
- Haroun
Shah, Head, Molecular Identification
Services, Department for Bioanalysis and
Horizon Technologies, Public Health England
For more details, please visit http://selectbiosciences.com/conferences/index.aspx?conf=CAMS2014
|
29-30 Oct 2014
|
Food Analysis Congress
Safety, Quality, Novel Technologies
Venue: Barcelona,
Spain
SELECTBIO’s inaugural Food Analysis
Congress aims to present the latest developments
in food analysis technologies, in response to the
increasing demand for rapid and efficient food
safety and quality testing.
Focus will be given to advances in both the
analysis of natural food allergens and toxins, as
well as contaminants introduced through processing
and packaging. Points for discussion will also
include the ongoing issue of food traceability and
efforts to reduce food fraud. Attending this event
will provide you with excellent opportunities for
networking with like minded peers, helping you to
find solutions and build collaborations.
Running alongside the conference will be an
exhibition covering the latest technological
advances and associated services from leading
solution providers within this field. Registered
delegates will also have access to the co-located
Clinical Applications of Mass Spectrometry
track, ensuring a cost effective trip.
For more
details, please visit http://selectbiosciences.com/conferences/index.aspx?conf=FAC2014
|
3-4 Dec 2014
|
Australian Lipids Meeting
Venue: University of Wollongong (Innovation
Campus), Wollongong, NSW, Australia
We are pleased to announce the 2nd Australian
Lipid Meeting, which will be held at the
University of Wollongong's Innovation Campus from
3-4 December, 2014.
While the first meeting focused on lipidomics we
have expanded the scope for the second meeting to
cover all aspects of lipid research. Planned
topics include:
- Imaging
- Botany
- Nutrition
- Health and Disease
- Technical Developments and Methodology
We look forward to seeing you in "The Gong".
Key Dates
Abstract submissions
- Open: 1 May, 2014
- Close: 31 July, 2014
- Acceptance notification: September 2014
Registration
- Open: 1 August, 2014
- Early bird: Closes 28 October, 2014
For
further details, please visit http://ihmri.uow.edu.au/research-program/australianlipidsmeeting/index.html
|
28 Jun to 2 Jul 2015
|
Metabolomics 2015: 11th
Annual International Conference of the
Metabolomics Society
The Official Annual Meeting of the Metabolomics
Society
Venue: San Francisco, USA
Details to follow.
Stay
abreast of the latest Metabolomics
Society news via the Twitter feed
on the front page of the website (http://www.metabolomicssociety.org).
Also
you
can
follow us on Twitter:
Metabolomics Society @MetabolomicsSoc
and Metabolomics journal
@Metabolomics.
And you can visit
us on Facebook.
Please
come back later for detailed
information about Metabolomics 2015 by
visiting
http://metabolomics2015.org.
|