The rising of
ambient-ionization mass spectrometry for metabolomics and
lipidomics applications
Feature
article contributed by Bindesh Shrestha1 and
Giuseppe Astarita1,2
1 Waters Corporation, Milford, MA 01757, USA
2 Department of Biochemistry and Molecular &
Cellular Biology, Georgetown University, Washington, DC 20007,
USA
Ambient ionization mass spectrometry (AIMS) consists of a
developing group of mass spectrometry (MS) ionization
techniques. With minimal sample preparation, these techniques
perform direct-sampling analyses at atmospheric pressure
(ambient conditions). Sampling and ion-generation take place in
concert, typically facilitating high analytical throughput.
Since the introduction of desorption electrospray ionization
(DESI) in 2004, at least 25 different AIMS ion sources have been
introduced, each with a unique combined approach to either
sampling or ionization. Recent advances in AIMS technology have
been discussed in detail.
1-3
In terms of the number of peer-reviewed publications, DESI has
led the pack among these AIMS techniques. A simple citation
search of “desorption electrospray ionization” returns more than
1100 peer-reviewed articles (Web of Science, Thomson Reuters,
New York, NY, accessed January 4, 2016). Articles published each
year from 2004 to 2015 are shown in
Figure
1 (top) with typical citations per article (bottom). Some
popular peer-reviewed journals that are publishing DESI-related
articles are tabulated in Table 1.
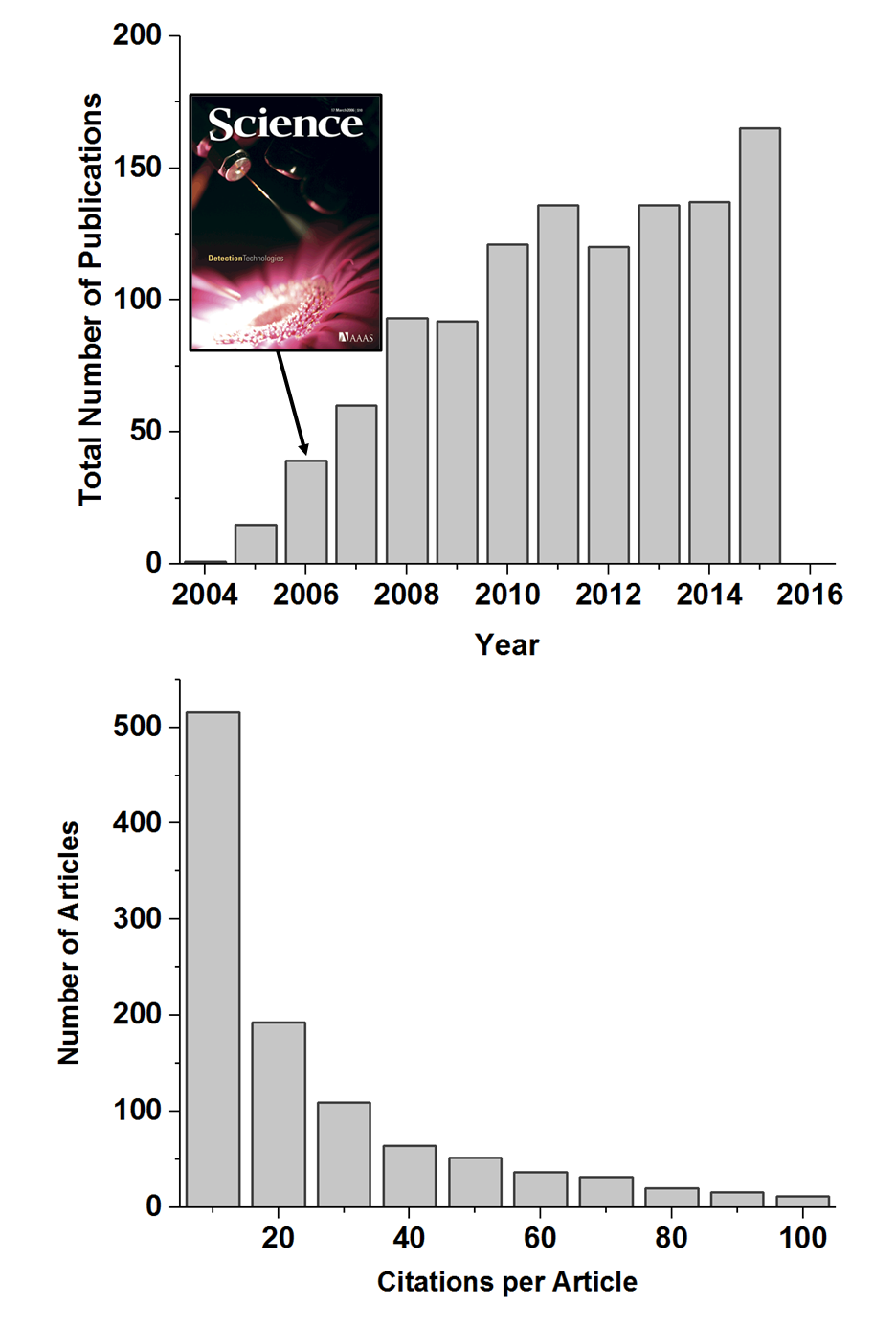 Figure 1.
Figure 1. Number of articles published per year, from
2004 to 2015, on the topic “desorption electrospray ionization”
(upper graph) and a trend of citation received per article
(lower graph). Based on Web of Science database (Thomson
Reuters, New York, NY).
Journal Name
|
Impact Factorα |
Number of Publications
|
Analytical Chemistry
|
5.636
|
203
|
Analyst
|
4.107
|
84
|
Journal of the American
Society for Mass Spectrometry
|
2.945
|
79
|
Rapid Communications in Mass
Spectrometry
|
2.253
|
76
|
Analytical and Bioanalytical
Chemistry
|
3.436
|
57
|
Journal of Mass Spectrometry
|
2.379
|
52
|
International Journal of Mass
Spectrometry
|
1.972
|
32
|
Analytica Chimica Acta
|
4.513
|
21
|
Analytical Methods
|
1.821
|
20
|
Proceedings of the National
Academy of Sciences
|
9.674
|
19
|
α Based on Journal Impact Factor (2014) from Thompson
Reuters, New York, NY.
Table 1. Top peer-reviewed journals with papers on the
topic of “desorption electrospray ionization”.
AIMS techniques have been employed to detect metabolites and
lipids directly from biofluids, mammalian tissue sections,
microbial colonies, cell cultures, etc.
4-6 AIMS
offers straightforward metabolomics and lipidomics workflows,
doing so with almost no reagent use and without time-consuming
sample extraction and other preparation steps. Samples are
analyzed outside the high-vacuum conditions of a mass
spectrometer, making AIMS conducive to the survival of live
specimens and thus creating the possibility of in vivo analysis.
Often only a sample-staging step, such as the sectioning of
tissue on microscope slides, is required. In small subsets of
AIMS, such as rapid evaporative ionization mass spectrometry
(REIMS),
7 sample staging is less critical or
unnecessary. In REIMS, for example, analytes can be extracted
directly from the surface of a sample, a tumor lesion from a
rodent, for example, or the skin of a fruit. Via long flexible
tubing, the samples are then transported to a mass spectrometer
for detection. Such an application demonstrates the ability of
AIMS to generate, in real time, molecular fingerprints that can
be used to discriminate among phenotypes.
In situ detection of lipids and metabolites from tissue is also
possible using other, conventional methods such as
matrix-assisted laser desorption/ionization (MALDI). However,
typical MALDI instrumentation requires the application of
organic molecules as a matrix and the introduction of the sample
into a vacuum. Like MALDI, AIMS techniques are constrained by
their incapacity to separate isobaric species and by ion
suppression effects. To circumnavigate this limitation, AIMS has
been used in combination with ion mobility separation, as a
post-ionization separation tool prior to MS detection, improving
the specificity of analyte identification and the
signal-to-noise ratio.
8-10
Figure
2 shows the power of ion mobility separation for
discriminating classes of biomolecules during molecular analysis
of mouse brain tissue using one of the AIMS techniques, laser
ablation electrospray ionization (LAESI).
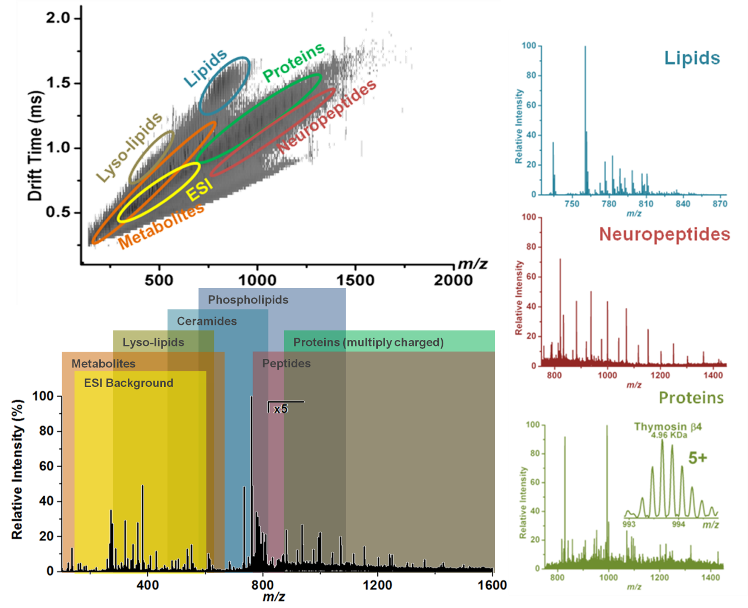 Figure 2.
Figure 2. The utility of ion mobility separation in
combination with ambient ionization mass spectrometry (AIMS).
Without ion mobility separation, many ions from different groups
of biomolecules can be difficult to separate. Mass spectra
discriminating just a group of molecules, such as lipids, can be
easily produced by selecting a defined
m/z and drift
time range (Adapted from Shrestha
et. al., Analytical
Chemistry 2014).
11
Molecular MS imaging is among the most remarkable applications
of AIMS. Briefly, in a typical MS-imaging experiment, a focused
laser beam or stream of charged droplets scans along a sample
section of surface tissue, obtaining a mass spectrum at each
defined pair of x and y coordinates. The intensities of detected
ions in those mass spectra can be plotted, to obtain maps of
corresponding metabolites and lipids in those tissue sections.
In terms of spatial resolution, DESI is capable of resolving
features in the tens of micrometers.
Figure
3 depicts an example of DESI image produced from a coronal
section of a mouse brain.
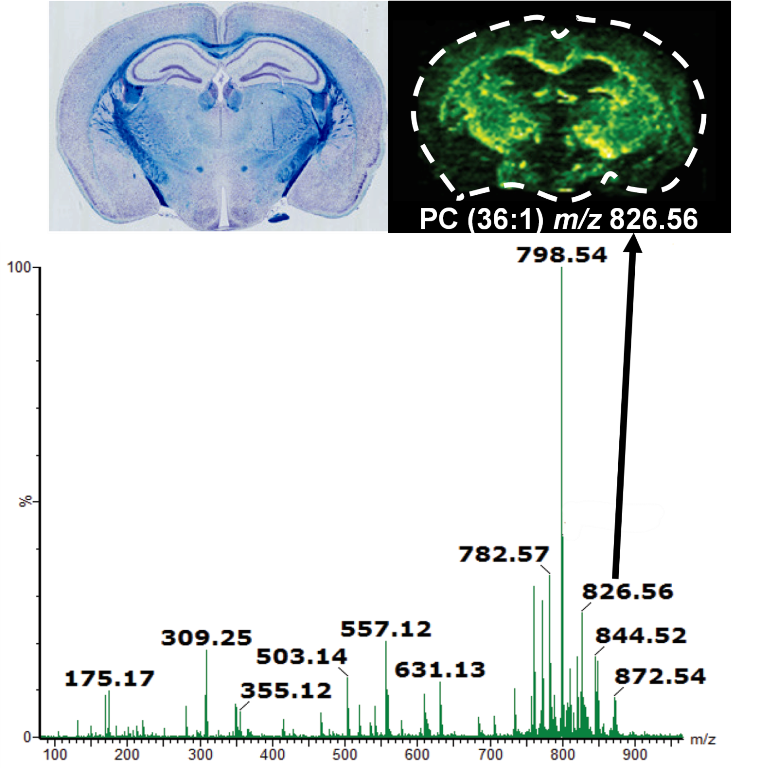 Figure 3.
Figure 3. Molecular image of phospholipids in a coronal
section of mouse brain obtained using DESI is presented
alongside an optical image of a concurrent section. A typical
positive mass spectrum of brain tissue using DESI is also shown
(Adapted from Waters Technical note# 720005297en).
Until recently, the majority of the AIMS-related applications
reported in the literature used home-built AIMS systems. These
home-built ion sources were typically coupled to the inlet of a
commercial mass spectrometer, usually in lieu of an electrospray
ion source. Several of the AIMS methods have been
commercialized, notable among them DESI, by Prosolia, LAESI, by
Protea, and direct analysis in real time (DART), by IonSense.
Usually those ion sources are retro-fitted onto commercial mass
spectrometers manufactured by Agilent, Bruker, Thermo, Waters,
JEOL, and others. In most cases, these bolted-on ion sources
require additional external software for instrument control and
data analysis workflow. Waters, however, has licensed the DESI
technology from Prosolia and also has acquired all assets
relating to REIMS technology from MediMass. Currently, both DESI
and REIMS are readily available as integrated options in both
the Waters SYNAPT and Xevo family of instruments. This
commercial solution, allowing for seamless instrument control
and data analysis, will encourage additional researchers to
adopt AIMS for their applications.
References
- Nemes, P., Vertes, A. TrAC Trends in Analytical
Chemistry 2012, 34, 22.
- Monge, M. E., Harris, G. A., Dwivedi, P., Fernández, F.
M. Chemical Reviews 2013, 113, 2269.
- Huang, M.-Z., Yuan, C.-H., Cheng, S.-C., Cho, Y.-T.,
Shiea, J. Annual review of analytical chemistry 2010,
3, 43.
- Wiseman, J. M., Ifa, D. R., Song, Q., Cooks, R. G. Angewandte
Chemie International Edition 2006, 45,
7188.
- Shrestha, B., Nemes, P., Nazarian, J., Hathout, Y.,
Hoffman, E. P., Vertes, A. Analyst 2010, 135,
751.
- Jackson, A. U., Tata, A., Wu, C., Perry, R. H., Haas, G.,
West, L., Cooks, R. G. Analyst 2009, 134,
867.
- Schäfer, K. C., Dénes, J., Albrecht, K., Szaniszló, T.,
Balog, J., Skoumal, R., Katona, M., Tóth, M., Balogh, L.,
Takáts, Z. Angewandte Chemie International Edition 2009,
48, 8240.
- Paglia, G., Kliman, M., Claude, E., Geromanos, S.,
Astarita, G. Analytical and bioanalytical chemistry
2015, 1.
- Stopka, S. A., Shrestha, B., Maréchal, É., Falconet, D.,
Vertes, A. Analyst 2014, 139, 5946.
- Inutan, E., Trimpin, S. Journal of the American
Society for Mass Spectrometry 2010, 21,
1260.
- Shrestha, B., Vertes, A. Analytical Chemistry
2014, 86, 4308.
Please note: If you know of any
metabolomics research programs, software, databases,
statistical methods, meetings, workshops, or training
sessions that we should feature in future issues of this
newsletter, please email Ian Forsythe at metabolomics.innovation@gmail.com.













