
MetaboNews
Issue 1 - August 2011
CONTENTS:
Online version of this newsletter:
http://www.metabonews.ca/Aug2011/MetaboNews_Aug2011.htm
 |
| MetaboNews Issue 1 - August 2011 |
|
CONTENTS: |
|
|
Welcome to the first issue of
MetaboNews, a monthly newsletter for the worldwide
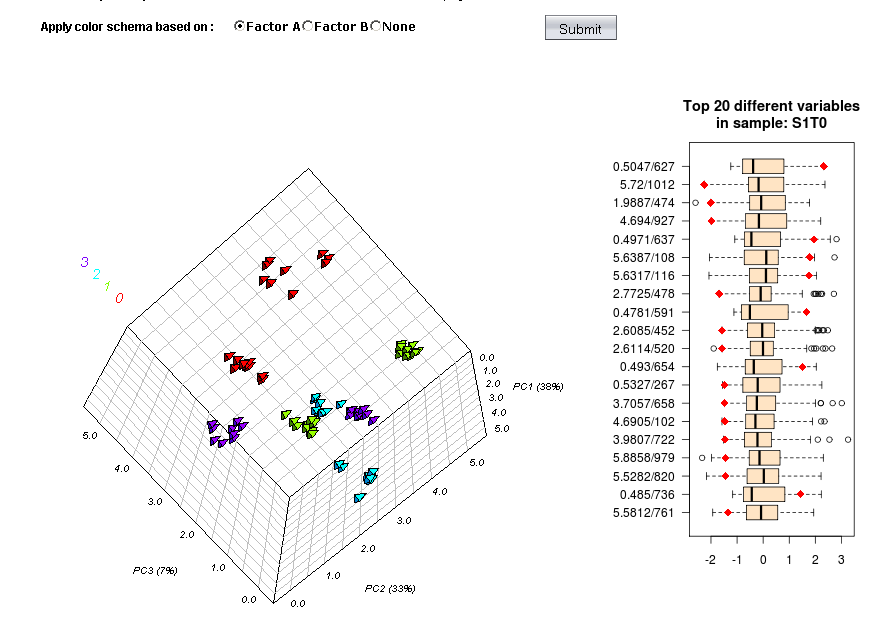
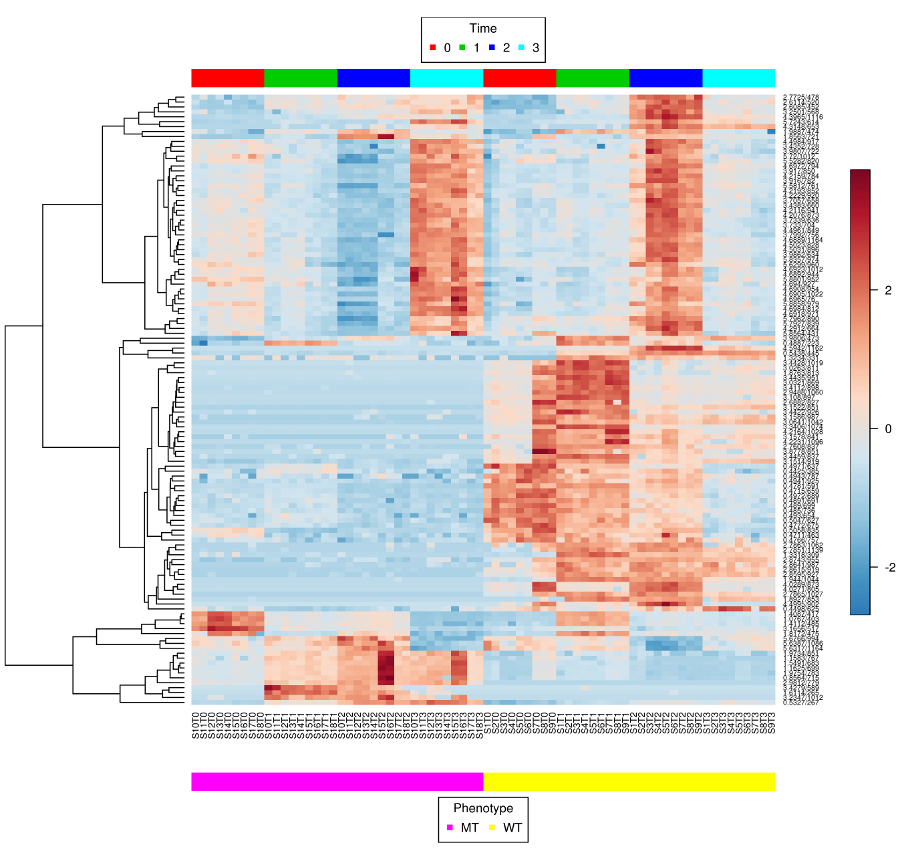
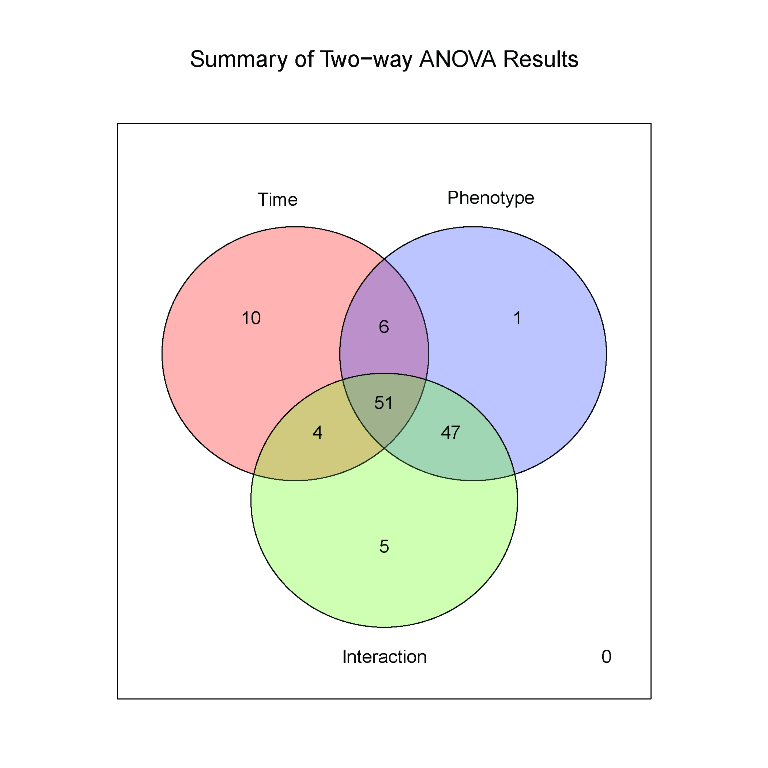
metabolomics community. In this issue we feature a Software
Spotlight article on MetATT, a web-based tool designed for
analyzing two-factor (including time-series) metabolomic data. This
newsletter is being produced by The Metabolomics Innovation
Centre (TMIC, http://www.metabolomicscentre.ca/) and is
intended to keep metabolomics researchers and other
professionals informed about new technologies, software,
databases, events, job postings, conferences, training
opportunities, interviews, publications, awards, and other
newsworthy items concerning metabolomics. We hope to provide
enough useful content to keep you interested and informed and
appreciate your feedback on how we can make this newsletter
better (metabolomics.innovation@gmail.com).
Note: Our subscriber list
is managed using Mailman, the GNU Mailing List Manager. To
subscribe or unsubscribe, please visit https://mail.cs.ualberta.ca/mailman/listinfo/MetaboNews
Back issues of this newsletter can be viewed from the newsletter archive (http://www.metabonews.ca/archive.html).
 |
1) Software Spotlight
|
| |
2) Biomarker Beacon
|
 |
3) Metabolomics Current Contents
|
 |
4) MetaboNews |
| Jul 2011 |
KEGG has gone commercial - Due to a lack of funding, one of Biology's most popular databases has moved to a subscription model. Minoru Kanehisa, a bioinformatics professor at Kyoto University, has led this project since its beginning in 1995. The KEGG website will continue to be freely available. However, academic and commercial users wanting access to KEGG's FTP site will now have to pay for access. Kanehisa and his colleagues have started a non-profit organization, NPO Bioinformatics Japan, as a means for raising funds to pay for KEGG's operations. Academic users will pay $2,000 per year for individual users and $5,000 per year for organizations. Kanehisa estimates that if they can get just 10% of the current FTP users (totalling 3,000 users per month), then they will have enough money to survive. A lot of work goes into making KEGG the high quality database that academic and commercial users have come to rely on during the last 15 plus years. As a result, KEGG employs a large team of 25 full-time staff and 5-10 part-time employees. To support KEGG and all of its efforts to date, consider a KEGG FTP subscription. |
| Jul 2011 |
Metabolomics Society meeting summary - The Seventh International Conference of the Metabolomics Society was held in Cairns, Australia, June 27-30, 2011, following relocation from Japan due to possible power shortages following tsunami. Despite the logistic challenges of such a last-minute move, the Australian hosts put together a great event with more than 140 posters and 80 speakers from around the world. The program was packed with presentations covering a wide variety of topics in metabolomics: everything from biomarker discovery, pathways and disease, plants, microbes and the environment, to pharmacology and nutrition. New technologies, fluxomics, bioinformatics, and systems biology were also hot topics. The emphasis on new capabilities in quantitative lipidomics was especially noteworthy. Another highlight was the presentation from Dr. Marasu Tomita, who is the director of the Institute for Advanced Biosciences, Keio University and was one of the original organizers for the conference. His talk emphasized the growing prominence of metabolomics in Japan, as reflected by the impressive metabolomics facilities in Tsuruoka city and their remarkable analytical capabilities. The south pacific, including Japan, Australia and New Zealand was generally very well represented at the conference, showing the growing global importance of metabolomics as an omics science. Japan will host the conference in 2014. Conference proceedings are available on the Metabolomics Society website at http://www.metabolomicssociety.org/news.html.7 |
 |
5)
Metabolomics Events
|
| 12-14 Sep 2011 |
CIC bioGUNE International Workshop on
Metabolomics CIC bioGUNE has prepared a very interesting scientific program in which leading academics speakers will tell us about the state-of-the-art in metabolomics including technological aspects related to the different platforms (RMN, GC and LC). The workshop will also cover recent breakthroughs in single-cell and high-throughput metabolomics approaches along with the applications that this technology has found in environmental and disease biomarker discovery areas. Importantly, the scientific program will be completed with poster sessions and presentations from equipment manufacturers. In summary the participants will have the opportunity to learn, meet and interact with experts in different metabolomics-related areas obtaining a deep and broad vision on this powerful technology. For more information,
please visit http://metabolomicsworkshop2011.cicbiogune.es/.
|
| 25-28 Sep 2011 |
Metabomeeting 2011 The MPF is also pleased to be organising Metabomeeting 2011 in conjunction with the inaugural Nordic Metabolomics meeting, which for this first meeting will be focusing on the latest developments/enhancements in data analysis and modelling. For more information,
please visit http://thempf.org/mpf_cms/index.php/conferencesworkshops/forthcoming-mpf-events/33-mm11.
|
| 30 Sep-4 Oct 2011 |
ASMS Asilomar Metabolomics meeting We will challenge ideas and concepts throughout the conference and have allocated ample time for discussions, e.g., during evening receptions. In addition to invited speakers, there will be poster sessions and oral contributions based on abstract submissions. We will focus on a range of hot topics, including: databases; metabolite/gene annotations; fluxes; compound identifications; metabolite imaging; secondary metabolism; metabolomics in human diseases; and quality control for high-throughput metabolomic quantifications. Download conference brochure For more information,
please visit http://www.asms.org/Conferences/AsilomarConference/GeneralInformation/tabid/99/Default.aspx.
|
| 8-9 Nov 2011 |
7th annual Advances in Metabolic Profiling The conference will be co-located with Mass Spec Europe. Registered delegates will have access to both meetings ensuring a very cost-effective trip. For more information,
please visit http://www.selectbiosciences.com/conferences/AMP2011/.
|
| 13-16 Nov 2011 |
International Conference on Natural Products
(ICNP2011) - Metabolomics
A
New Frontier in Natural Products Science The conference is being organised by the Universiti Putra Malaysia and Malaysian Natural Products Society. The conference will be held at Palm Garden Hotel, IOI Resort, PUTRAJAYA from 13th -16th November 2011. For more information,
please visit http://www.icnp2011.upm.edu.my/index.html.
|
| 25-28 June 2012 |
METABOLOMICS 2012: Breakthroughs in plant,
microbial and human biology, clinical and
nutritional research, and biomarker discovery Local Organisers: Dan Bearden, Rick Beger, Rima Kaddurah-Daouk, Lloyd W. Sumner, Don Robertson, Padma Maruvada and Donna Kimball. For more information,
please visit http://www.metabolomics2012.org.
|
Please note: If you know of any metabolomics lectures, meetings, workshops, or training sessions that we should feature in future issues of this newsletter, please email Ian Forsythe (metabolomics.innovation@gmail.com).
 |
6) Metabolomics Jobs |
This is a resource for advertising positions in metabolomics.
If you have a job you would like posted in this newsletter,
please email Ian Forsythe (metabolomics.innovation@gmail.com).
Job postings will be carried for a maximum of 4
issues (8 weeks) unless the position is filled prior to that
date.
Jobs
Offered
| Job Title | Employer | Location | Date Posted | Source |
|---|---|---|---|---|
| Postdoctoral
Research Associate position to analyze plant
composition |
Iowa State University |
Ames, Iowa |
25-Aug-2011 |
Metabolomics
Society Jobs |
| Postdoctoral
Fellow in Metabolomics |
Duke University |
Durham, NC |
20-Aug-2011 |
Metabolomics
Society
Jobs |
| Scientist,
Proteomics/Metabolomics |
Merck Sharp & Dohme (MSD) Singapore |
Singapore |
20-Aug-2011 |
JobsDB.com |
| Post-Doctoral
Fellow: Biomarkers of Disease in Dairy Cows |
University of Alberta |
Edmonton, AB |
3-Aug-2011 |
University
of Alberta |
|
Ian J.
Forsythe, M.Sc.
MetaboNewsEditor Department of Computing Science
University of Alberta 221 Athabasca Hall Edmonton, AB, T6G 2E8, Canada Email: metabolomics.innovation@gmail.com Website: http://www.metabonews.ca |
This newsletter is produced by The
Metabolomics Innovation Centre (TMIC, http://www.metabolomicscentre.ca/) for
the benefit of the worldwide metabolomics community.
|