GC×GC-TOFMS strategy for combined quantitative and untargeted
profiling of polar metabolites
Feature article contributed by Tuulia Hyötyläinen
and Matej Orešič, VTT
Technical Research Centre of Finland, Espoo, FIN-02044 VTT,
Finland
Metabolomics aims for comprehensive characterization of
metabolites and their pathways in biological systems. Ideally, any
analytical strategy for metabolomics would thus allow for broad as
well as sensitive coverage of a metabolome across several chemical
and functional classes of metabolites. While such approaches are
already possible today, they usually come at a cost. The so-called
untargeted metabolic profiling methods are usually qualitative or
semi-quantitative, thus not enabling determination of absolute
concentrations of the metabolites measured. This presents at least
two challenges: (1) it is difficult to compare the results across
different experiments and platforms, and (2) knowledge of absolute
concentrations may be important for the interpretation of the
data. On the other hand, targeted approaches which cover only a
pre-selected set of metabolites are usually more limited in
metabolite coverage and do not allow for the discovery of novel,
important metabolites in the specific experimental setting.
Over the past years we have been establishing a metabolomics
platform which combines the targeted as well as the untargeted
approach, based on two-dimensional gas chromatography coupled to
time-of-flight mass spectrometry (GC×GC-TOFMS) [
1].
Compared to traditional GC-MS approaches, the sensitivity of
GC×GC-TOFMS is typically 10-40 times higher and the separation
efficiency is up to 20-fold better. Moreover, the quality of mass
spectra obtained with GC×GC-TOFMS is much better than those
obtained with GC-MS, and thus, identification is more reliable.
Method development of GC-MS methods (including GC×GC-TOFMS) is
straightforward and much easier than e.g., LC-MS. Also,
quantitation is more reliable, because matrix effects in GC-MS are
generally not a critical issue. An additional advantage of all
GC-based methods is the repeatable characteristics of retention,
particularly when nonpolar stationary phases are used. This makes
it possible to use universal retention indices, which are highly
useful in the identification of unknown compounds, in combination
with mass spectral detection. Large spectral libraries for
GC(×GC)-MS, combined with retention index data is a valuable tool
in the identification of unknown compounds. Even if a particular
MS spectrum is not present in the spectral database, the
fragmentation pattern can be used to obtain information about the
compound class of a metabolite [
1].
Over 1000 metabolites can be detected by GC×GC-TOFMS in a single
sample run, covering a functionally wide range of small molecules
such as amino acids, fatty acids, sterols, carboxylic acids,
sugars, alcohols, and amines. Of interest to biomedical
researchers and microbiologists, the platform can also detect many
metabolites produced by gut microbiota [
2].
Despite being first introduced to the metabolomics community
already several years ago [
3],
the GC×GC-TOFMS-based metabolic profiling approach has not yet
been widely adopted in metabolomics. There have been three major
challenges concerning the application of the platform: (1)
derivatization of polar metabolites is required, (2) the
qualitative nature of the data when a global screening approach is
adopted, and (3) difficulties with processing large amounts of
data, given their multi-dimensional nature.
In our research, we have aimed to overcome the limitations of
GC×GC-TOFMS (
Figure
1). As usual in GC-MS-based metabolomics methods,
derivatization of metabolites is performed in a sequential,
automated manner using an autosampler. In order to enable
semi-quantitative and quantitative analysis in parallel, internal
standards for selected metabolites to be quantified are added
prior to extraction. The selected metabolites, typically about 30
in number, are then quantified using the external calibration
curves and by manual checks of peak integration. This step is time
consuming and is performed by an expert analytical chemist.
Additional internal standards are utilized also for the global
profiling of all other metabolites, where the data undergoes
automated data processing. Data for these metabolites is
normalized using the internal standards, thus enabling
semi-quantitative analysis [
1].
In order to facilitate global metabolic profiling by GC×GC-TOFMS,
we also developed an open source software package
Guineu for
automated processing of GC×GC-TOFMS data [
1].
Guineu takes the processed data (peak list) from vendor software
for each sample, then performs additional steps such as alignment
of the data, normalization, data filtering, utilization of
retention indices in the verification of identification as well as
automated group-type identification of the compounds.
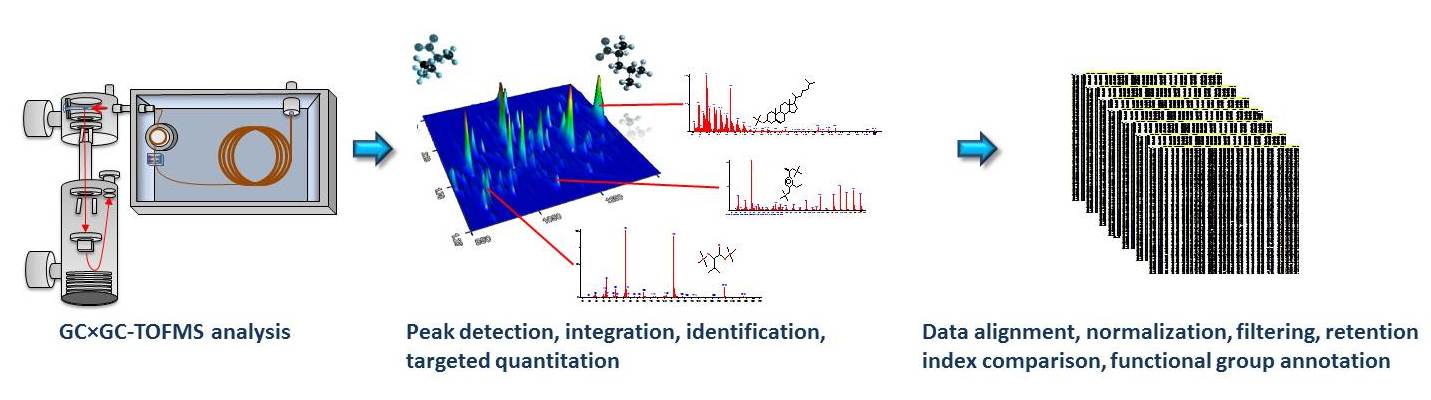 Figure 1.
Figure 1. Typical GC×GC-TOFMS metabolic profiling
workflow.
Although the platform is repeatable over extended periods of time
[
1],
metabolic profiling by GC×GC-TOFMS is unlikely to be applied in
studies requiring analyses of large numbers of samples such as in
epidemiological studies. The throughput is still rather low for
such studies, requiring typically about 45 minutes for a single
sample analysis. Given its extensive coverage of the metabolome,
metabolic profiling by GC×GC-TOFMS is particularly suited for
tackling problems where the metabolome is still poorly
characterized (e.g., in the brain, or at the host-microbial
interface) or where the discovery of novel biomarkers is sought.
For example, we found GC×GC-TOFMS to be a powerful analytical
strategy in our studies of diseases of the central nervous system.
In an Alzheimer’s disease (AD) study, an untargeted approach by
GC×GC-TOFMS allowed us to identify a plasma metabolite
2,4-dihydroxybutyric acid which is associated with later
progression to AD [
4].
In a follow-up study, we also found that this metabolite is a
strong predictor of AD when measured in cerebrospinal fluid (CSF)
(unpublished data), thus suggesting the detected changes in plasma
reflect the changes in brain metabolism in early AD. Recently,
GC×GC-TOFMS was also applied to characterize patient-matched
plasma and CSF samples [
5],
thus providing a valuable resource on the composition of the CSF
metabolome and how it compares to the plasma metabolome.
In conclusion, metabolic profiling by GC×GC-TOFMS is a powerful
approach for comprehensive characterization of small polar
metabolites, allowing for both quantitative and semi-quantitative
analysis in parallel. Its unique strengths make GC×GC-TOFMS a
complementary platform to other more established analytical
strategies for metabolomics such as LC-MS(/MS) and NMR.
References
[1] S. Castillo, I. Mattila, J. Miettinen, M. Orešič,
T. Hyötyläinen, Data analysis tool for comprehensive
two-dimensional gas chromatography-time of flight mass
spectrometry, Anal. Chem. 83, 3058–3067 (2011). [PMID:
21434611]
[2] A.-M. Aura, I. Mattila, T. Hyötyläinen, P. Gopalacharyulu,
C. Bounsaythip, M. Orešič, K.-M. Oksman-Caldentey, Drug
metabolome of the Simvastatin formed by human intestinal
microbiota in vitro, Mol. BioSyst. 7, 437-446 (2011). [PMID:
21060933]
[3] W. Welthagen, R.A. Shellie, J. Spranger, M. Ristow, R.
Zimmermann, O. Fiehn, Comprehensive two-dimensional gas
chromatography time-of-flight mass spectrometry (GC-GC-TOF) for
high resolution metabolomics: biomarker discovery on spleen
tissue extracts of obese NZO compared to lean C57BL/6 mice,
Metabolomics 1, 65-73 (2005).
[4] M. Orešič, T. Hyötyläinen, S.-K. Herukka, M. Sysi-Aho, I.
Mattila, T. Seppänan-Laakso, V. Julkunen, P. V. Gopalacharyulu,
M. Hallikainen, J. Koikkalainen, M. Kivipelto, S. Helisalmi, J.
Lötjönen, H. Soininen, Metabolome in progression to Alzheimer’s
disease, Transl. Psychiatry 1, e57 (2011). [PMID:
22832349]
[5] M. Hartonen, I. Mattila, A.-L. Ruskeepää, M. Orešič, T.
Hyötyläinen, Characterization of cerebrospinal fluid by
comprehensive two-dimensional gas chromatography coupled to
time-of-flight mass spectrometry, J. Chromatogr. A 1293, 142-149
(2013). [PMID:
23642768]
Please
note: If you know of any
metabolomics research programs, software, databases,
statistical methods, meetings, workshops, or training
sessions that we should feature in future issues of this
newsletter, please email Ian Forsythe at metabolomics.innovation@gmail.com.








