The Golm Metabolome Database (GMD)
Jan Hummel, Kenny Billiau, Alexander Erban, Dirk Walther,
Joachim Kopka
Max Planck Institute for Molecular Plant Physiology,
Potsdam-Golm, Germany
{hummel|erban|walther|kopka}@mpimp-golm.mpg.de
The Golm Metabolome Database (
http://gmd.mpimp-golm.mpg.de;
Kopka et al., 2005, Schauer et al., 2005, Hummel et al., 2007) has
been developed and is hosted by the Max Planck Institute for
Molecular Plant Physiology (Golm, Germany), a pioneer in applying
metabolomic studies to plant research. The GMD harbors information
on reference compounds and compounds detected in biological
samples, respective mass spectra and retention index information
for compound annotation in complex mixtures (Schauer et al.,
2005), and metabolite level measurements derived primarily from
plant material and probed with gas chromatography/mass
spectrometry (GC/MS) technology. It currently comprises
information on 1,167 reference substances, 1,676 metabolites,
2,588 electron impact ionization mass spectra associated with
known compounds and 3,145 spectra for which the compound is not
yet identified.
The GMD—a resource for compound annotation by matching to GC/MS
reference spectra and retention indices
The GMD can be queried for particular metabolites, reference
compounds and respective chemical derivatives searching with text,
sum formula, molecular weight, functional chemical groups, KEGG
identifiers, and other properties. All compounds contained in the
GMD are displayed with a broad array of chemical and physical
property information and links to external data resources (
Figure
1). Most notably, the GMD permits mass spectra-based queries
(
Figure
2) thereby facilitating the annotation process of
user-obtained GC/MS spectra.
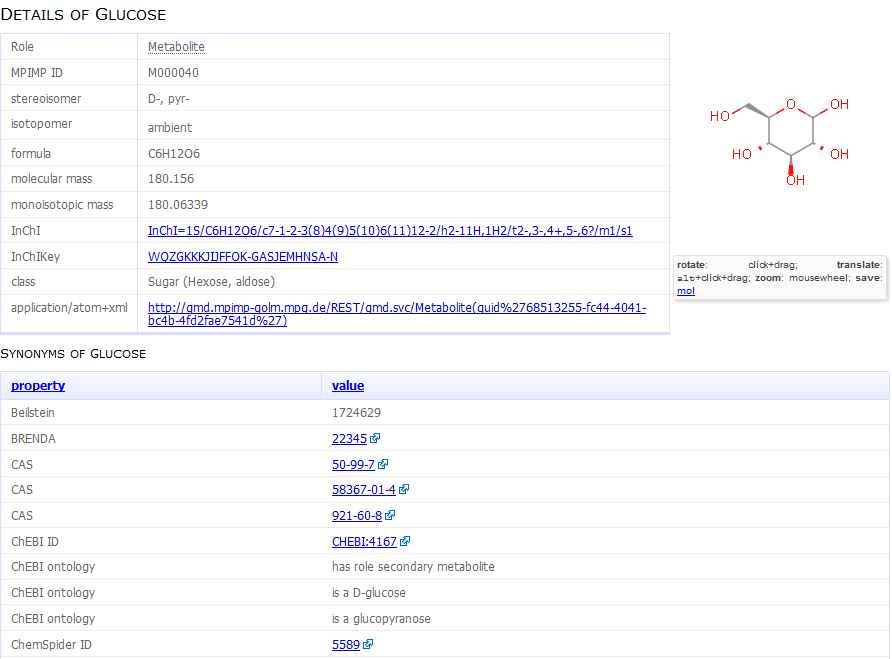 Figure 1.
Figure 1. Example of a metabolite report card (excerpt)
providing detailed chemical information on particular compounds
contained in the GMD.
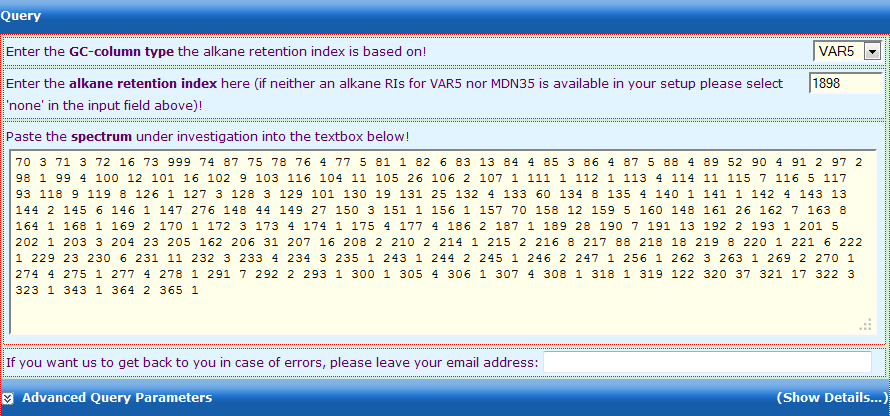 Figure 2.
Figure 2. Input form for querying the GMD with a mass
spectrum. The query can be performed with or without RI
constraints. Two standard RI-systems are supported (Strehmel et
al., 2008).
Applying several scoring methods for spectra comparison, matching
compounds with similar mass spectra will be ranked and returned in
tabular format (
Figure
3). Graphical support facilitates the manual evaluation of
the automated identification result and the selection of the best
matching compound.
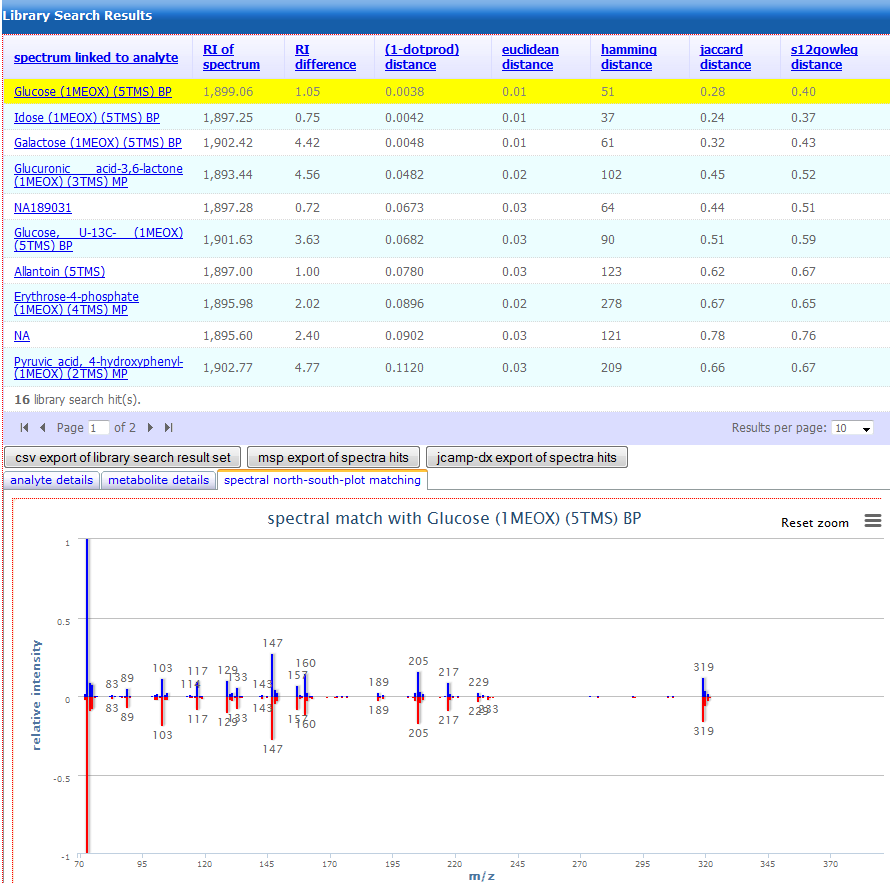 Figure 3.
Figure 3. Example of a result page obtained from a
mass spectrum query. Compounds with similar spectra compared to
the user-submitted query are tabulated along with various sortable
match-scores and a graphical display of the selected mass spectral
hit.
Automated characterization of mass spectra—annotating “unknown”
metabolites
Based on the comprehensive compound and associated spectra
information content of the GMD, a automated,
machine-learning-based annotation of “unknown” spectra has been
implemented (Hummel et al., 2010, Hummel et al., 2013). Employing
decision trees trained in a rigorous cross-validation setting,
mass spectra can be annotated for the presence or absence of a
number of 21 different functional groups (
Figure
4) that frequently occur in known metabolites, e.g., amino-,
alcohol-, or carboxylic acid-moieties. Thereby, even spectra for
which no matching reference compound or respective metabolite
spectrum can be found in the database, a basic classification of
the “unknown” compound can be generated allowing, for example, to
identify the likely candidate compounds for subsequent
identification steps.
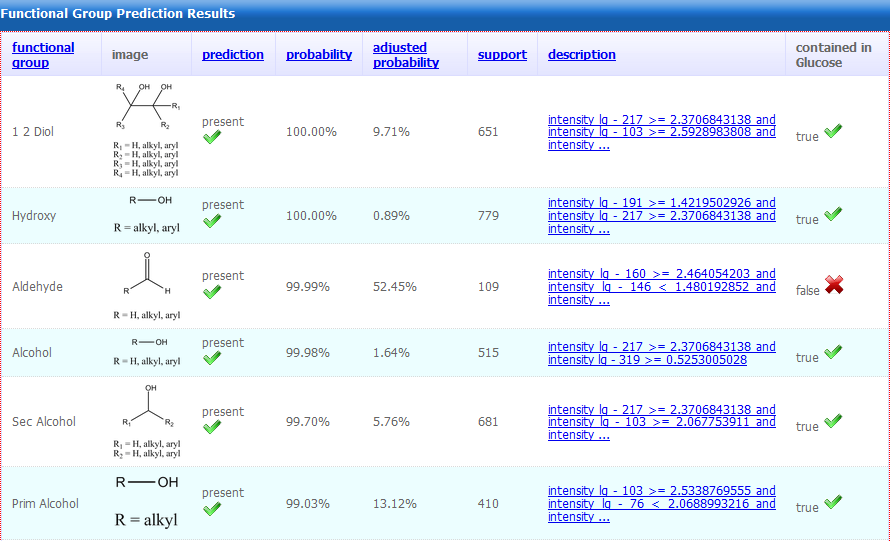 Figure 4.
Figure 4. Mass spectra are annotated for the
presence or absence of 21 different characteristic functional
groups providing assistance in the annotation of compound spectra
of unknown identity. Prediction scores and classification rules
are provided (Hummel et al., 2010).
From compound-centric to cross-experiment data views
Recently, the development focus of the GMD has shifted from
providing compound and spectra-centric towards integrated views of
compound sets associated with a particular experimental series,
and beyond this experiment-centric view, offering means for the
comparative analysis of metabolite profiles across different
experiments. Currently, 14 experiments have already been
incorporated into the GMD. Various graphics-supported analytical
displays facilitate the analysis of compound profiles across
different experiments and within single experimental datasets (
Figure
5). Compound profiles can also be queried with hypothetical
compound level constellations, such as, queries that require
metabolite A to be present, metabolite B absent, and metabolite C
at twice the level of compound A. In the long term this
functionality aims at permitting “experiment” queries which answer
questions, such as, under which other conditions do metabolites
exhibit similar levels as in a user’s experiment. With the rapidly
growing number of experimental datasets, this facility is expected
to quickly gain appeal.
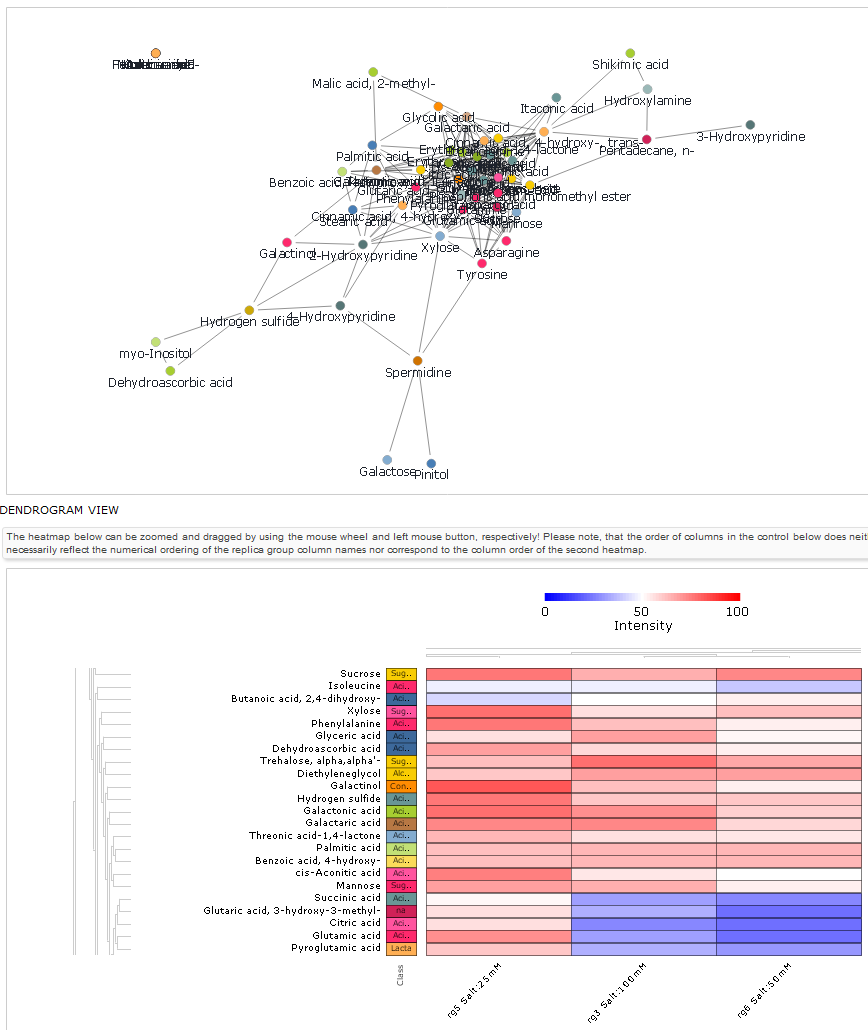
Figure 5. The GMD permits analyses of metabolite
level information across all samples associated with a particular
experiment. Similar metabolite profiles are identified via
correlation methods and displayed graphically as network graphs
(upper panel) and employing hierarchical clustering methods
producing heat map views of the metabolite level data (lower
panel).
Inferface to external software systems via web services
The GMD offers a wide array of software interface functionalities
allowing the integration of GMD-functionality, e.g., automated
classification of mass spectra, into third-party software
solutions.
Terms of Use
Access to the online contents and services of the GMD is granted
to all users without any limitations pursuant to the regulations
stipulated in the GMD website. Commercial licenses for off-line
use are available upon request.
References
- Hummel, J., Selbig, J., Walther, D. and Kopka, J. (2007) The
Golm Metabolome Database: a database for GC-MS based
metabolite profiling. In Nielsen, J. and Jewett, M.C. (eds), Metabolomics.
Springer-Verlag, Berlin, Heidelberg, New York, 75-96.
- Hummel, J., Strehmel, N., Bölling, C., Schmidt, S., Walther,
D., Kopka, J. (2013) Mass spectral search and analysis using
the Golm Metabolome Database. In Weckwerth, W., and Kahl, G.
(eds) The Handbook of Plant Metabolomics. Wiley-VCH
Verlag, Weinheim, 321-343.
- Hummel, J., Strehmel, N., Selbig, J., Walther, D., Kopka, J.
(2010) Decision tree supported substructure prediction of
metabolites from GC-MS profiles. Metabolomics, 6, 322-333.
- Kopka, J., Schauer, N., Krueger, S., Birkemeyer, C., Usadel,
B., Bergmuller, E., Dormann, P., Weckwerth, W., Gibon, Y.,
Stitt, M., Willmitzer, L., Fernie, A.R. and Steinhauser, D.
(2005) GMD@CSB.DB: The Golm Metabolome Database,
Bioinformatics, 21, 1635-1638.
- Schauer, N., Steinhauser, D., Strelkov, S., Schomburg, D.,
Allison, G., Moritz, T., Lundgren, K., Roessner-Tunali, U.,
Forbes, M.G., Willmitzer, L., Fernie, A.R., Kopka, J. (2005)
GC-MS libraries for the rapid identification of metabolites in
complex biological samples, FEBS Letters, 579, 1332-1337.
- Strehmel, N., Hummel, J., Erban, A., Strassburg, K., Kopka,
J. (2008) Retention index thresholds for compound matching in
GC-MS metabolite profiling, Journal of Chromatography B, 871,
182-190.
Acknowledgements
The development of the GMD was supported by grants from the
Deutsche Forschungsgemeinschaft (DFG) and the EU (FP-7) as part of
the COSMOS initiative
http://www.cosmos-fp7.eu/.
Please
note: If you know of any
metabolomics research programs, software, databases,
statistical methods, meetings, workshops, or training
sessions that we should feature in future issues of this
newsletter, please email Ian Forsythe at metabolomics.innovation@gmail.com.















