
MetaboNews
Issue 5 - December 2011
CONTENTS:
Online version of this newsletter:
http://www.metabonews.ca/Dec2011/MetaboNews_Dec2011.htm
 |
| MetaboNews Issue 5 - December 2011 |
|
CONTENTS: |
|
|
Welcome to the fifth issue of
MetaboNews, a monthly newsletter for the worldwide
metabolomics community. In this month's issue, we feature a Software Spotlight
article on Agilent Technologies' new GeneSpring-Mass Profiler
Professional (MPP) 12.0. This
newsletter is being produced by The Metabolomics Innovation
Centre (TMIC, http://www.metabolomicscentre.ca/), and is
intended to keep metabolomics researchers and other
professionals informed about new technologies, software,
databases, events, job postings, conferences, training
opportunities, interviews, publications, awards, and other
newsworthy items concerning metabolomics. We hope to provide
enough useful content to keep you interested and informed and
appreciate your feedback on how we can make this newsletter
better (metabolomics.innovation@gmail.com).
Note: Our
subscriber list is managed using Mailman, the GNU Mailing List
Manager. To subscribe or unsubscribe, please visit https://mail.cs.ualberta.ca/mailman/listinfo/MetaboNews
Current and back issues of this newsletter can be viewed from the newsletter archive (http://www.metabonews.ca/archive.html).
 |
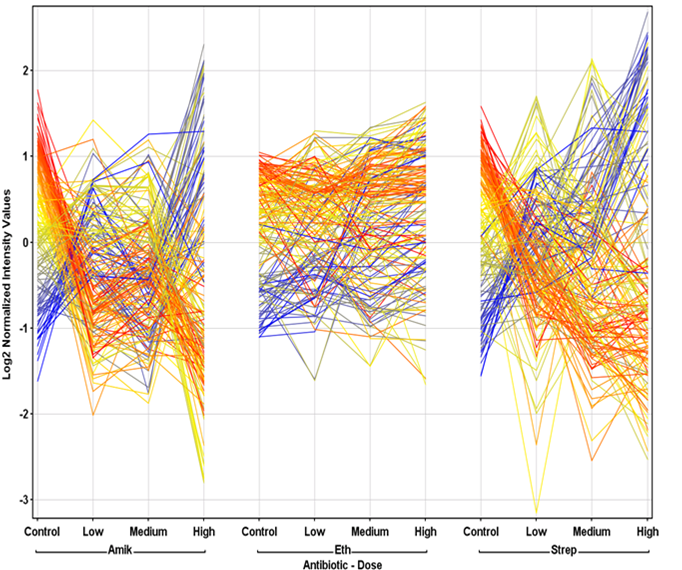
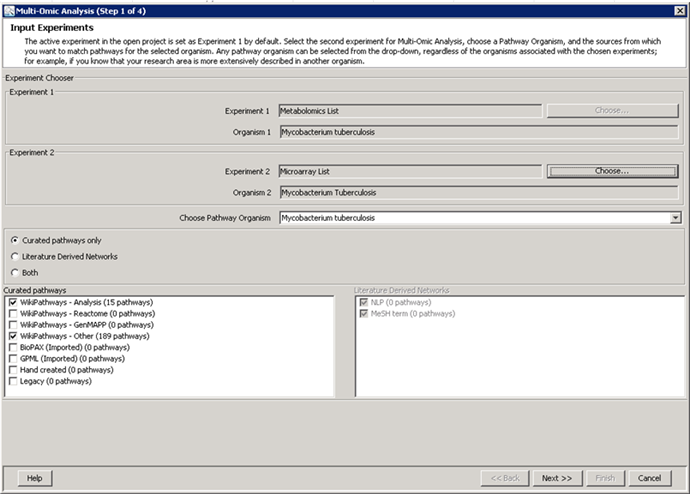
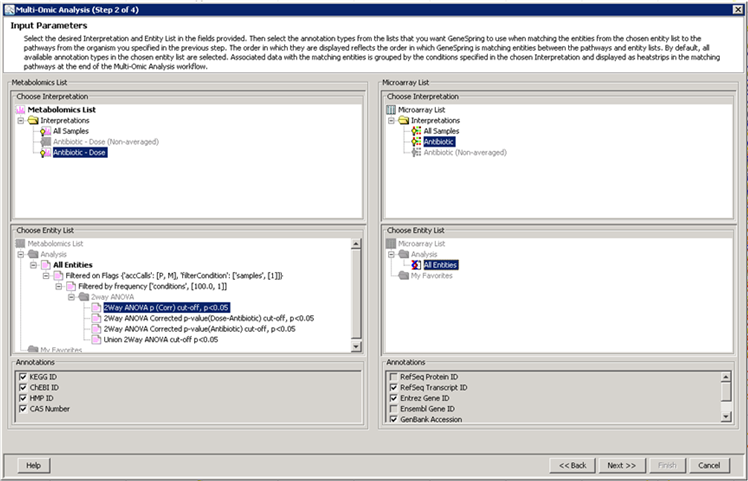
1) Software Spotlight
|
 |
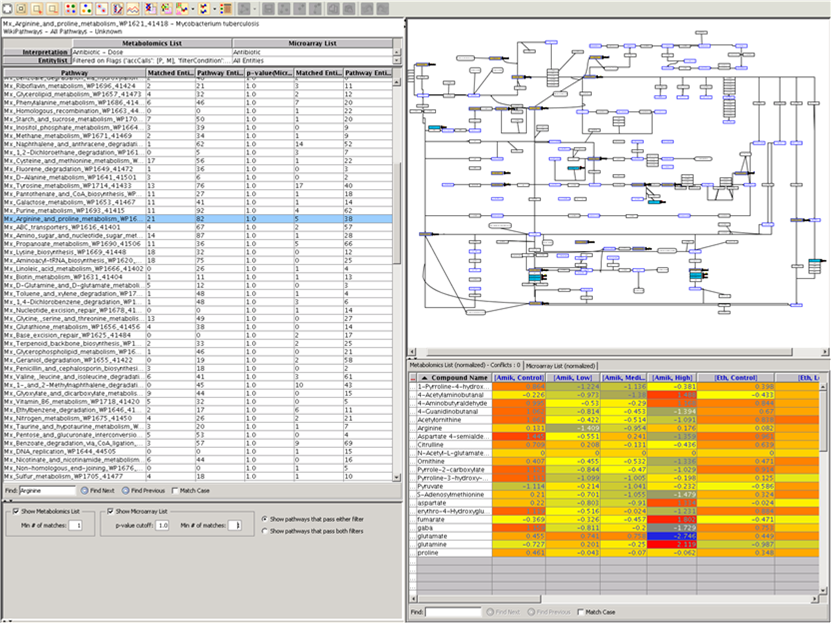
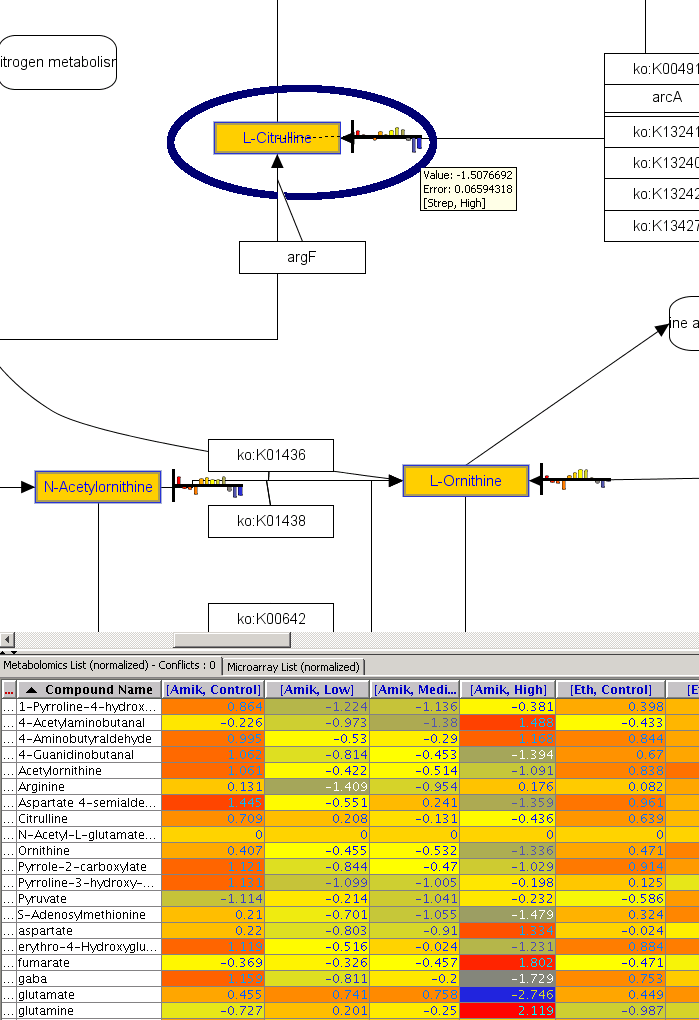
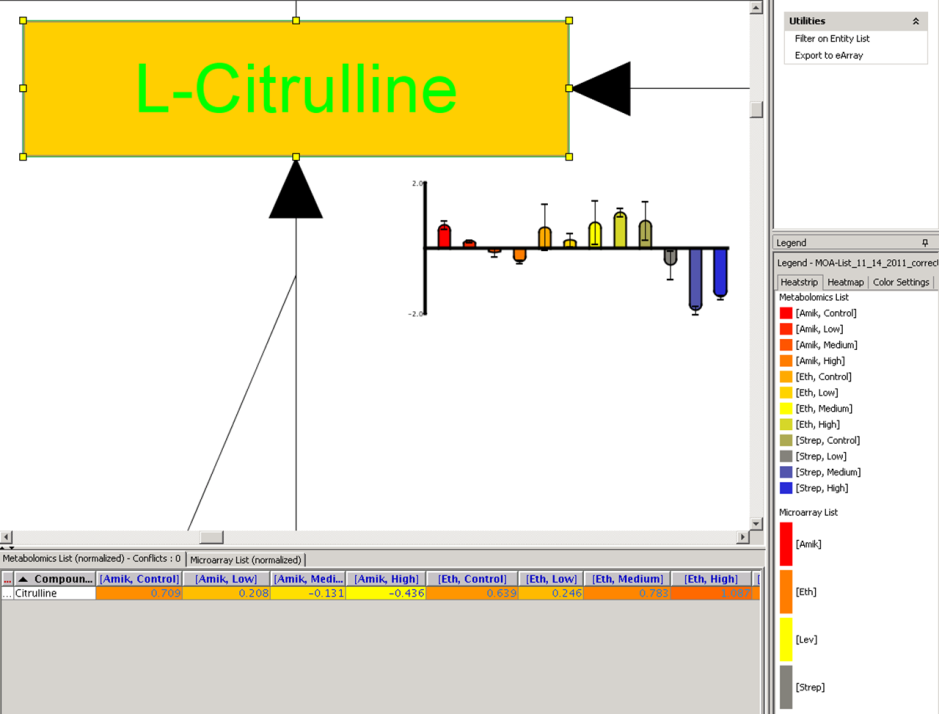
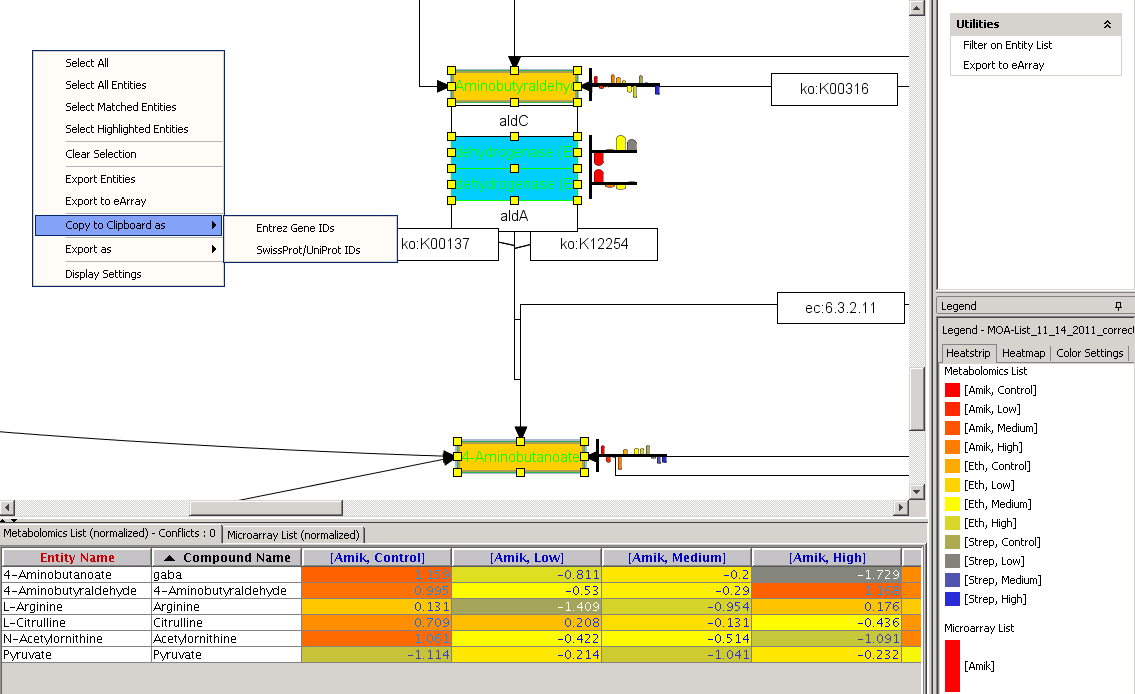
| |
2) Biomarker Beacon
|
 |
3) Metabolomics Current Contents
|
 |
4) MetaboNews |
| 15 Dec 2011 |
Metabolomx test detects
lung cancer from breath - Metabolomx, a
diagnostic company focused on the detection of the
metabolomics signature of cancer from exhaled
breath, today announces publication of results
from the first clinical study demonstrating a
breath test that can both detect lung cancer and
differentiate between types of lung cancer in
humans. This seminal study, conducted at the
Cleveland Clinic and led by Dr. Peter Mazzone,
used Metabolomx' first-generation colorimetric
sensor array, and reported accuracy exceeding 80%
in lung cancer detection, comparable to
computerized tomography (CT) scan. Further, the
study found that Metabolomx' first-generation
colorimetric sensor array could identify the
subtype of lung cancer (small cell versus
adenocarcinoma versus squamous cell) with accuracy
approaching 90%.
The availability of a low-cost, non-invasive metabolomic breath signature for lung cancer is particularly timely given the recently announced results of the National Cancer Institute's National Lung Screening Trial (NLST) calling for wider CT screening of high-risk citizens. The breath signature, which reports active tumor metabolism, is thought to provide complementary information to CT, potentially helping clinicians distinguish benign from malignant lung nodules. The sensor detects the unique pattern of volatile organic compounds (the "metabolic biosignature") present in exhaled breath. The article, "Exhaled Breath Analysis with a Colorimetric Sensor Array for the Identification and Characterization of Lung Cancer," is appearing in the current, online issue of the Journal of Thoracic Oncology (JTO), the official Journal of the International Association for the Study of Lung Cancer. Paper: Mazzone PJ et al., Exhaled breath analysis with a colorimetric sensor array for the identification and characterization of lung cancer. J Thorac Oncol. 2012 Jan;7(1):137-42. [PMID: 22071780] |
| 14 Dec 2011 |
Biochemical fingerprint
in the blood predicts Alzheimer's disease - A study led by
Research Professor Matej Oresic from VTT suggests
that Alzheimer´s disease is preceded by a
molecular signature indicative of hypoxia and
up-regulated pentose phosphate pathway. This
indicator can be analysed as a simple biochemical
assay from a serum sample months or even years
before the first symptoms of the disease occur. In
a healthcare setting, the application of such an
assay could therefore complement the
neurocognitive assessment by the medical doctor
and could be applied to identify the at-risk
patients in need of further comprehensive
follow-up.
Alzheimer’s disease (AD) is a growing challenge to the health care systems and economies of developed countries with millions of patients suffering from this disease and increasing numbers of new cases diagnosed annually with the increasing aging of populations. The progression of Alzheimer’s disease (AD) is gradual, with the subclinical stage of illness believed to span several decades. The pre-dementia stage, also termed mild cognitive impairment (MCI), is characterised by subtle symptoms that may affect complex daily activities. MCI is considered as a transition phase between normal aging and AD. MCI confers an increased risk of developing AD, although the state is heterogeneous with several possible outcomes, including even improvement back to normal cognition. What are the molecular changes and processes which define those MCI patients who are at high risk of developing AD? The teams led by Matej Orešič from VTT and Hilkka Soininen from the University of Eastern Finland set out to address this question, and the results were published on 13th Dec. 2011 in Translational Psychiatry. The team used metabolomics, a high-throughput method for detecting small metabolites, to produce profiles of the serum metabolites associated with progression to AD. Serum samples were collected at baseline when the patients were diagnosed with AD, MCI, or identified as healthy controls. 52 out of 143 MCI patients progressed to AD during the follow-up period of 27 months on average. A molecular signature comprising three metabolites measured at baseline was derived which was predictive of progression to AD. Furthermore, analysis of data in the context of metabolic pathways revealed that pentose phosphate pathway was associated with progression to AD, also implicating the role of hypoxia and oxidative stress as early disease processes. The unique study setting allowed the researchers to identify the patients diagnosed with MCI at baseline who later progressed to AD and to derive the molecular signature which can identify such patients at baseline. Though there is no current therapy to prevent AD, early disease detection is vital both for delaying the onset of the disease through pharmacological treatment and/or lifestyle changes and for assessing the efficacy of potential AD therapeutic agents. The elucidation of early metabolic pathways associated with progression to Alzheimer’s disease may also help in identifying new therapeutic avenues. This study was supported by the project “From patient data to personalised healthcare in Alzheimer's disease” (PredictAD) which was supported by the European Commission under the 7th Framework Programme. Paper: M. Orešič, T. Hyötyläinen, S. -K. Currant, Sysi-Aho, M., I. Mattila, T. Seppänen-Laakso, V. Julkunen, PV Gopalacharyulu, M. Hallikainen, J. Koikkalainen, M. Stone Field, S. Heli Salmi, J. Lötjönen, H. Soininen, Thurs metabolome in the progression of Alzheimer's disease, Translational Psychiatry (2011) 1, E57; doi: 10.1038/tp.2011.55. http://www.nature.com/tp/journal/v1/n12/full/tp201155a.html |
| 8 Dec 2011 |
Metabolon Study Reveals
Mechanisms of High Dosage Pentamethyl-6-Chromanol
(PMCol) Toxicity in Liver Damage and Dysfunction -
Metabolon, Inc., the leader in metabolomics, biomarker
discovery and biochemical analysis, announced today
the publication of "Toxicogenomics and Metabolomics of
Pentamethylchromanol (PMCol)-Induced Hepatotoxicity"
in the journal Toxicological
Sciences. The study was conducted by the
Biosciences Division of SRI International in
conjunction with Metabolon scientists.
The study integrated classical toxicology, toxicogenomics, and metabolomic approaches to determine the mechanism of toxicity and identify sensitive early markers of target organ injury by PMCol. Metabolomic evaluation of the liver and plasma samples found dose- and time-dependent effects, including depletion of total glutathione and glutathione conjugates, decreased methionine and cofactors FAD and NAD(P)+, and increased S-adenosylhomocysteine, cysteine, and cystine. Chronic exposure to high doses of PMCol induced liver damage and dysfunction that is likely due to two factors: the direct inhibition of glutathione synthesis and the modification of drug metabolism pathways. Paper: Parman T et al., Toxicogenomics and metabolomics of pentamethylchromanol (PMCol)-induced hepatotoxicity. Toxicol Sci. 2011 Dec;124(2):487-501. Epub 2011 Sep 13. [PMID: 21920950] Source: MarketWatch |
 |
5)
Metabolomics Events
|
| 1 Feb 2012 |
Recent Advances and Future Directions in
Applied Metabolomics
Speakers include:
For more information, please visit http://www.coebp.ls.manchester.ac.uk/documents/AppliedMetabolomicsFeb1stflyer_000.pdf.
|
| 3 Feb 2012 |
Cancer Metabolomics: Elucidating the
Biochemical Programs that Support Cancer Initiation
and Progression Call for Poster Abstracts A poster session will be held and a selected number of presenters will be asked to give brief oral presentations. The deadline for abstract submission is Friday, January 27, 2012. For abstract instructions, send an email to CancerMetabolomics@nyas.org with the words "Abstract Information" in the subject line. Instructions will be forwarded automatically. For questions, please call 212.298.8618. For more information, please visit http://www.nyas.org/Events/Detail.aspx?cid=7fbbd804-93df-4a78-8127-8210ad9f3496.
|
| 20-22 Feb 2012 |
International Conference and Exhibition on
Metabolomics & Systems Biology Metabolomics-2012 will serve as a catalyst for the advances in the study of Metabolomics & Systems Biology by connecting scientists within and across disciplines at sessions and exhibition held at the venue, creates an environment conducive to information exchange, generation of new ideas, and acceleration of applications that benefit Research in Metabolomics & Systems Biology. Conference Highlights the following topics:
For more information, please visit http://omicsonline.org/metabolomics2012/.
|
| 9-10 Mar 2012 |
Second International Congress of
Translational Research in Human Nutrition:
Integrative approaches in nutrition research Details to follow: http://thempf.org/mpf_cms/index.php/conferencesworkshops/other-forthcoming-events
|
| 17-20 Apr 2012 |
analytica Conference 2012 In various symposiums, leading scientists from all over the world report on the latest developments, current trends and visions of the future. Analytic, diagnostic, biochemical and molecular biological methods and procedures are discussed here. On the last occasion, 140 well-known experts gave talks in 23 different thematic symposiums. Main subject emphases/highlights of the analytica Conference 2010
For more information, please visit http://www.analytica.de/en/Home/congress/conference-program.
|
| 7-8 May 2012 |
LIPID MAPS Annual Meeting 2012: Lipidomics
Impact on Cell Biology, Metabolomics and
Translational Medicine The meeting program tentatively features the following six sessions:
For more information, please visit http://www.lipidmaps.org/meetings/2012annual/
|
| 20-24 May 2012 |
60th ASMS Conference on Mass Spectrometry and
Allied Topics Important Dates: Jan 9, 2012:
Conference registration and lodging opens online
Feb 3, 2012: Deadline for submission of abstracts Apr 14, 2012: Conference program online Apr 30, 2012: Deadline for advance registration For more information, please visit http://www.asms.org/Conferences/AnnualConference/GeneralInformation/tabid/127/Default.aspx.
|
| 21-23 May 2012 |
Les 6èmes Journées
Scientifiques du Réseau Français de
Métabolomique et Fluxomique (JS 6 RFMF)
For more information, please visit https://www.bordeaux.inra.fr/ifr103/reseau_metabolome/document/meetings/courrier_preinscription_6JSRFMF_final.rtf
|
| 25-28 June 2012 |
METABOLOMICS 2012: Breakthroughs in plant,
microbial and human biology, clinical and
nutritional research, and biomarker discovery Local Organisers: Dan Bearden, Rick Beger, Rima Kaddurah-Daouk, Lloyd W. Sumner, Don Robertson, Padma Maruvada and Donna Kimball. For more information, please visit http://www.metabolomics2012.org.
|
| 10 July 2012 |
Merck Deep Dive: Translational Technologies
for Basic and Clinical Scientists For more information, please visit http://www.cvent.com/events/merck-deep-dive-molecular-biomarkers-protemics-genomics-metabolomics-/event-summary-be463e01b8974ad99e3d84af4be50bef.aspx.
|
 |
6) Metabolomics Jobs |
This is a resource for advertising positions in metabolomics.
If you have a job you would like posted in this newsletter,
please email Ian Forsythe (metabolomics.innovation@gmail.com).
Job postings will be carried for a maximum of 4
issues (8 weeks) unless the position is filled prior to that
date.
Jobs
Offered
| Job Title | Employer | Location | Date Posted | Source |
|---|---|---|---|---|
| Global Marketing Manager - LC/MS Business | Agilent Technologies | Santa Clara, CA, USA | 28-Dec-2011 |
LinkedIn Jobs |
| Research Associate, Mathematical Modeling - Applied Mathematics & Modeling | Merck | Greater Philadelphia Area, USA | 16-Dec-2011 |
LinkedIn Jobs |
| Associate Director - Bioinformatics | Genentech | United States |
16-Dec-2011 |
LinkedIn Jobs |
| Postdoctoral researcher in environmental metabolomics | University of Birmingham | Birmingham, UK | 15-Dec-2011 |
Metabolomics Society Jobs |
| Post-doctoral position (18 months) - Cancer, Environment and Metabolomics | LABERCA |
Nantes, France |
13-Dec-2011 |
Réseau Français de Métabolomique et Fluxomique |
| Postdoctoral Research Associate in Functional Genomics | Oak Ridge National Laboratory | Oak Ridge, Tennessee, USA |
9-Dec-2011 |
LinkedIn Jobs |
| Stable Isotopes Scientist | Nestlé Research Center | Lausanne, Switzerland | 1-Dec-2011 |
Réseau
Français de Métabolomique et Fluxomique |
| Research
Engineer position in Biostatistics with skills in OMICS techniques |
L.C.H., the French horse doping control laboratory | Verrières le Buisson, France |
1-Dec-2011 | Réseau Français de Métabolomique et Fluxomique |
| Three
Assistant/Associate Professorships in Metabolomics and Computational Biology |
University of Birmingham, UK | Birmingham, UK | 29-Nov-2011 |
Metabolomics Society Jobs |
|
Ian J.
Forsythe, M.Sc.
MetaboNewsEditor Department of Computing Science
University of Alberta 221 Athabasca Hall Edmonton, AB, T6G 2E8, Canada Email: metabolomics.innovation@gmail.com Website: http://www.metabonews.ca LinkedIn: http://ca.linkedin.com/in/iforsythe |
This newsletter is
produced by The Metabolomics Innovation Centre (TMIC, http://www.metabolomicscentre.ca/)
for the benefit of the worldwide metabolomics
community.
|