The
Birmingham Metabolomics Training Centre – Current
requirements and new opportunities for training in
metabolomics
Ralf
J. M. Weber1, Catherine L. Winder1,
Lee D. Larcombe2, Mark R. Viant1 and
Warwick B. Dunn1
1Birmingham Metabolomics Training Centre,
School of Biosciences, University of Birmingham,
Edgbaston, Birmingham B15 2TT, UK
2ELIXIR-UK, Department of Physiology, Anatomy
& Genetics, University of Oxford, Oxford OX1 3PT, UK
Training needs have been identified for our expanding
metabolomics community
The metabolomics community is growing at a rapid rate.
This is shown by the increasing number of publications during
the last 12 years (32 and 2414 publications listed on PubMed
in 2003 and 2014, respectively) and the increasing number of
large-scale metabolomics centres being developed globally (for
example, the MRC-NIHR National Phenome Centre in the UK and
the six NIH-funded Regional Comprehensive Metabolomics
Resource Cores in the USA). In any scientific discipline that
is growing rapidly there is a considerable requirement for
introductory and specialised training in the relevant
laboratory and computational workflows. Indeed, the lack of
skilled professionals can inhibit the growth of the
metabolomics community [1].
Recently, we conducted an international survey to determine
the training needs of the metabolomics community. The survey
was conducted in association with the international
Metabolomics Society (http://metabolomicssociety.org/)
and ELIXIR-UK (http://elixir-uk.org/).
The Metabolomics Society was established to promote the growth
of this relatively new scientific field, of which training
early-career scientists and scientists who have not applied
metabolomics previously is a core objective. ELIXIR-UK is the
BBSRC/MRC/NERC funded UK node within the ELIXIR infrastructure
in the European Union and focuses on bioinformatics training
provision in partnership with other European ELIXIR Nodes.
ELIXIR-UK has prioritized 5 critical areas of UK training
need. Metabolomics is one of those priorities and this
collaborative effort will be key to engaging the community and
addressing the emerging skills gap. Our international survey
had the objectives to determine the training courses currently
available, areas where training is required and the delivery
mechanisms preferred by the community. The results and
recommendations were recently summarised in the journal Metabolomics
[2] and all results are available on the Metabolomics
Society website (http://metabolomicssociety.org/publications/training-needs-in-metabolomics).
There were 202 responses to the survey. The results indicated
that the current training courses are not sufficient to meet
the growing training needs of the international metabolomics
community. To allow the metabolomics community to expand
through professionally trained scientists we need to develop
appropriate training courses in both theoretical aspects of
metabolomics as well as hands-on training in laboratory and
computational skills. The requirements in laboratory and
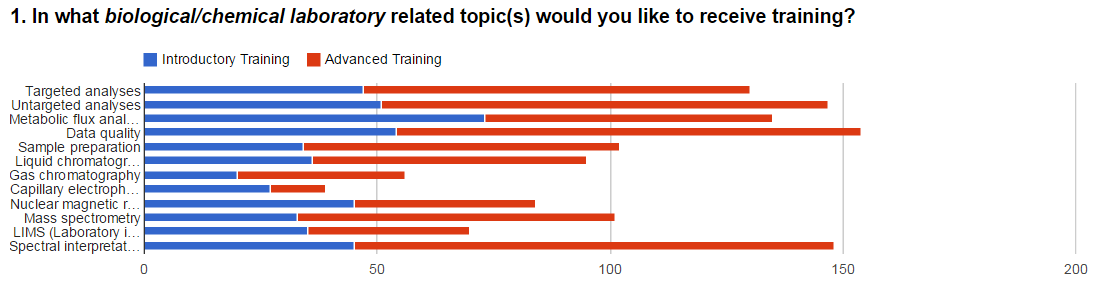
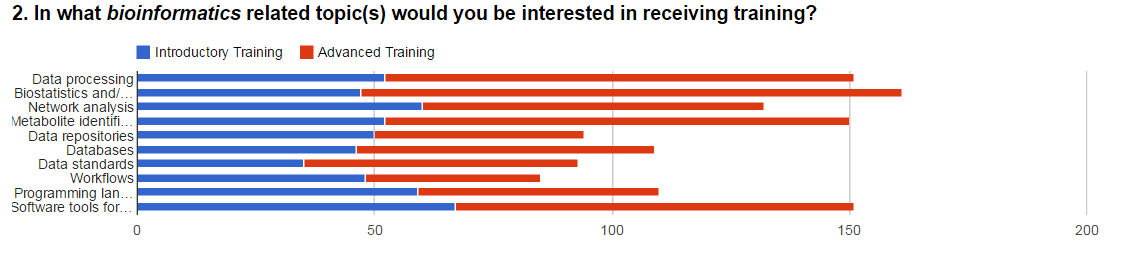
bioinformatics courses at beginners and advanced level are
shown below in Figure 1. A Wordle is also shown in Figure 1
for the different databases applied.
A
B
C

Figure 1. Survey results showing the responses to
questions focused on training requirements in (A)
chemical/analytical laboratories, (B) bioinformatics, and (C)
databases applied in metabolomics.
The key recommendations arising from this survey are to
[2],
- Develop a series of face-to-face (f2f) and e-learning
training courses to fill the knowledge gaps in analytical
metabolomics and bioinformatics.
- Create new funding opportunities to build national and
international networks of trainers to both develop and
deliver training programs, thereby maximising existing
investments of the funders into research projects and
facilities.
- Improve the advertising and accessibility of both existing
and new training courses, for example via the ELIXIR TeSS
portal (https://tess.elixir-uk.org/)
and Metabolomics Society network.
- To initiate discussions within the international
metabolomics community to consider a formal accreditation of
training programs in order to achieve high training
standards, harmonisation across courses and the on-going
redevelopment of courses as technologies evolve.
The newly launched Birmingham Metabolomics Training Centre

The requirement for a greater number of training courses in
different areas of metabolomics has been highlighted in the
training survey discussed above. The University of Birmingham,
UK, has internationally recognised metabolomics expertise and
hosts national facilities to support both clinical (Phenome
Centre-Birmingham) and environmental (NERC Biomolecular Analysis
Facility - Birmingham) science, applying both mass spectrometry
and NMR spectroscopy as well as data processing and analysis.
This wealth of expertise has been used to establish the
Birmingham Metabolomics Training Centre (BMTC,
http://www.birmingham.ac.uk/facilities/metabolomics-training-centre/index.aspx)
as a new centre of excellence for metabolomics training across
multiple topics within metabolomics including both analytical
and computational methods. The courses developed to date as well
as those being planned are targeted to fill known gaps in
training as identified by the training survey. A combination of
hands-on, face-to-face courses and online courses will be
developed that incorporate different and innovative teaching
methods. The face-to-face workshops will include lectures,
laboratory practicals, and computational sessions with
significant hands-on training utilising state-of-the-art
analytical instruments and software. We will collaborate with
analytical instrument and software providers to provide training
in new cutting-edge developments. These courses will be marketed
to the metabolomics community as a whole or to specialised
communities, for example courses aimed at environmental
scientists or clinical scientists.
To provide an optimal environment for hands-on training, the
number of trainees per session will be strictly limited (Dr
Elizabeth Want, International Phenome Training Centre, personal
communication). To provide training to larger audiences in a
distributed approach to allow the trainee to select when they
perform the training each week, online training courses need to
be developed. These courses should provide interaction between
educators and learners and allow forum discussions between
learners to which educators can contribute. In 2013, the
University of Birmingham joined a consortium to deliver online
training or
MOOCs (Massive Open Online Courses) via the
UK-based FutureLearn platform (
https://www.futurelearn.com/).
MOOCs are free to register for, deliver learning in small
segments, offer flexibility in time and study, and consequently
are available to a wide audience. The MOOC detailed below was
the first worldwide to focus on metabolomics.
MOOC: Metabolomics: Understanding metabolism in the 21
st
century (register at
https://www.futurelearn.com/courses/metabolomics)
It ran for four weeks in October 2015 and attracted 2262
learners. The course was targeted towards MSc and PhD students
but we envisaged that it would also provide a valuable
introduction to metabolomics for scientists at any stage in
their careers. The learning community actually included both
scientists and non-scientists from school pupils to retired
people. A key aspect of MOOCs is the social learning (2883
comments were posted across the course) and we (the educators)
greatly enjoyed interacting with the learning community and
answering their questions. The overriding opinions from the
learners were that they found the course to be very enjoyable
and challenging, and appreciated the introduction to this
fascinating subject. So we plan to continue introducing this
online community to metabolomics and will hopefully run the
course again next year.
In conclusion, the metabolomics community have strongly
indicated the requirement for more training capacity and have
highlighted the areas where new training courses are required as
well as the preferred types of training delivery. To assist in
supporting the growth of the international metabolomics
community the Birmingham Metabolomics Training Centre has been
established to deliver both face-to-face and online training
courses at both introductory and advanced levels. We hope the
training survey will lead to the development of further training
courses internationally to meet the needs of the metabolomics
community.
Acknowledgements
We thank the BBSRC, MRC and NERC for funding of the ELIXIR-UK
node (BB/L005077/1), and thank the Metabolomics Society for
support in undertaking the metabolomics training questionnaire.
References
[1] Metabolomics Market By Technique, Application and by
Indications – Global Forecasts to 2019, Marketsandmarkets.com,
report July 2019.
[2] Ralf J. M. Weber, Catherine L. Winder, Lee D. Larcombe,
Warwick B. Dunn, & Mark R. Viant. Training Needs in
Metabolomics, (2015). Metabolomics, 11:784–786
Please note: If you know of any
metabolomics research programs, software, databases,
statistical methods, meetings, workshops, or training
sessions that we should feature in future issues of this
newsletter, please email Ian Forsythe at metabolomics.innovation@gmail.com.












