
TMIC and the Metabolomics Society
Issue 66 - February 2017
CONTENTS:
Online version of this newsletter:
http://www.metabonews.ca/Feb2017/MetaboNews_Feb2017.htm

 |
| Published
in partnership between TMIC and the Metabolomics Society Issue 66 - February 2017 |
|
CONTENTS: |
|
|
 |
| TMIC
Services |
 |
Metabolomics Society News |

|
|
 |
| |
Metabolomics Spotlight
|
| |
MetaboInterview
|
 |
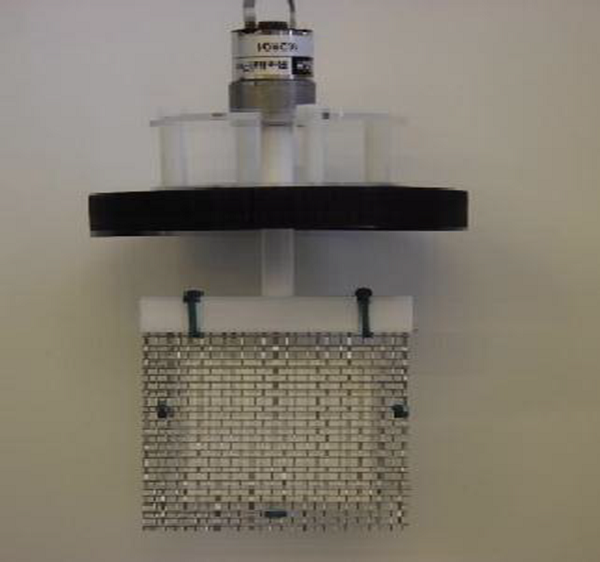
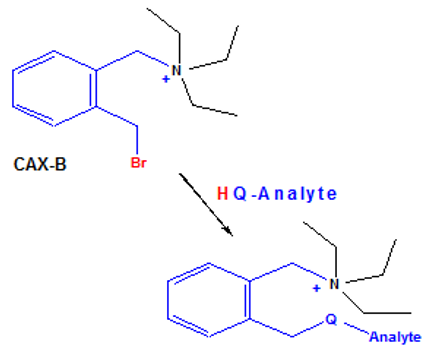
Biography Roger W. Giese, PhD is a Professor of Chemistry and Biomedical Science, in the Department of Pharmaceutical Sciences at Northeastern University in Boston. Dr. Giese is interested in measuring the human exposome by mass spectrometry, and in increasing analytical specificity and sensitivity in metabolomics. |
|
|
 |
|
|
 |
 |
Metabolomics Current
Contents
|
 |
MetaboNews |
| 9 Jan 2017 |
Diagnosing,
detecting, and predicting
diseases just got six
million times easier
The Canada Foundation for Innovation (CFI) just gave a $6 million vote of confidence to medical research in the burgeoning field of metabolomics with this morning’s announcement of funding for Canada’s national metabolomics laboratory, the Metabolomics Innovation Centre (TMIC), hosted at the University of Alberta. Metabolomics provides scientists with a powerfully precise route to study drug metabolism, gene-environment interactions, and biochemical processes. “Metabolomics uses some of the latest techniques in analytical chemistry and computational biology to measure the chemicals that living organisms need—such as amino acids and vitamins—as well as the chemicals that living organisms are exposed to, such as drugs, pesticides, and pollutants,” says David Wishart, professor in the University of Alberta Faculty of Science and lead of TMIC. “It is shining a new light on how chemicals affect human, animal, and plant health and leading to some really innovative approaches for diagnosing and predicting disease.” Source: University of Alberta Faculty of Science |
Metabolomics Events
|
| 8-9 Feb 2017 |
Computational Environmental Metabolomics Course Venue: Birmingham Metabolomics Training Centre, University of Birmingham, Birmingham, UK This 2-day NERC funded Advanced Training Short Course will provide a theoretical overview and hands-on training in the processing and analysis of metabolomics data. You will study specific case studies from the environmental sciences, learning the step-by-step processes that are applied in the data analysis pipeline. The course will be delivered using a combination of lectures and computer workshops, and time will be dedicated to answering questions and discussing your projects. Topics include:
|
| 10-14 Feb 2017 |
Phenome 2017 Venue: Hilton El Conquistador Resort, Tucson, Arizona, USA The Phenome 2017 conference is an important step in the development of a path toward training and collaboration among disciplines that are poised to address critical social issues and to generate greater understanding about plants and climate change. This first conference will bring together a multidisciplinary community comprising plant biologists, ecologists, engineers, agronomists, and computer scientists. Please share this flyer with your members of your department and encourage your students to register to attend the meeting. Highly relevant abstracts submitted by November 1 may be considered to give a talk during one of the sessions. Please visit www.phenome2017.org to register and learn more about the meeting. |
| 13-17 Feb 2017 |
EMBO Practical Course on Metabolomics Bioinformatics for Life Scientists Venue: European Bioinformatics Institute (EMBL-EBI) - Training Room 2 - Wellcome Genome Campus, Hinxton, Cambridge, CB10 1SD, UK Application opens: Monday August 08 2016 Application deadline: Friday November 11 2016 Participation: Open application with selection Contact: Maria Bacadare Goitia Registration fee: £350 Overview This course will provide an overview of key issues that affect metabolomics studies, handling datasets and procedures for the analysis of metabolomics data using bioinformatics tools. It will be delivered using a mixture of lectures, computer-based practical sessions and interactive discussions. The course will provide a platform for discussion of the key questions and challenges in the field of metabolomics, from study design to metabolite identification. Audience This course is aimed at PhD students, post-docs and researchers with at least one year’s experience in the field of metabolomics who are seeking to improve their skills in metabolomics data analysis. Participants ideally must have working experience using R (including a basic understanding of the syntax and ability to manipulate objects). For more information, please visit https://www.ebi.ac.uk/training/events/2017/embo-practical-course-metabolomics-bioinformatics-life-scientists-3. |
| 20 Feb to 17 Mar 2017 |
Metabolomics Data Processing and Data Analysis Online Course An online course by the Birmingham Metabolomics Training Center, University of Birmingham, UK hosted by Futurelearn Venue: Birmingham Metabolomics Training Centre, University of Birmingham, Birmingham, UK This four-week online course will explore the tools and approaches that are used to process and analyse metabolomics data, we will investigate the challenges that are typically encountered in the analysis of metabolomics data and provide solutions to overcome these problems. The course will be delivered using a combination of short videos, articles, discussions, and online workshops with step-by-step instructions and test data sets. We will provide quizzes, polls and peer review exercises each week, so that you can review your learning throughout the course. The material will be delivered over a four week period, with an estimated learning time of four hours per week. If you do not have time to complete the course during the 4-week period you will retain access to the course material to revisit, as you are able. Registration fee: Early-bird £200, Standard £220 For further information and registration details, please visit http://www.birmingham.ac.uk/facilities/metabolomics-training-centre/courses/Metabolomics-Data-Processing-and-Data-Analysis.aspx or contact bmtc@contacts.bham.ac.uk. |
| 13-15 Mar 2017 |
CPSA Metabolomics 2017 Venue: The University of Florida Clinical & Translational Science Institute, Gainesville, Florida, USA 3rd Annual Metabolomics Symposium on Clinical and Pharmaceutical Solutions through Analysis (CPSA Metabolomics 2017) "… differential nutrient utilization patterns can identify subsets of cancers with distinct tendencies for potential pharmacological intervention." - Allen, EL; Ulanet, DB; Pirman, D; et al. Cell Rep. 2016, Oct 11; 17; 876-890. Technologies and Methodologies for Drug Discovery Our Discussion Leader, John Janiszewski of Pfizer, has organized a provocative session to help delineate the role of novel technologies and methods for drug discovery that range from biomarker validation to fluxomics to computational approaches. Please make plans for engaging discussions and visit the recently updated program agenda to review our line-up of sessions and exciting events at this year's annual meeting. Call for Papers Abstracts are being accepted for the 3rd Annual Metabolomics Symposium on Clinical and Pharmaceutical Solutions through Analysis (CPSA Metabolomics 2017)! Click on Call for Papers for author instructions. Submit your abstract by February 28, 2018. Registration Registration is open! Click on the CPSA Metabolomics 2017 registration link and register today! Travel & Accommodations Prepare now for your Travel & Accommodations for CPSA Metabolomics 2017. Make your hotel reservations at the Hilton University of Florida Conference Center. Reserve your room on-line or call the hotel directly (352-371-3600). For more information, visit http://www.cpsa-metabolomics.com.
|
| 13-17 Mar 2017 |
Hands-on LC-MS for Metabolic Phenotyping Venue: Imperial College London, South Kensington, London, UK Description: This week long course aims to cover how to perform a metabolic profiling experiment, from start to finish. It covers study design, sample preparation, the use of mass spectrometry for global profiling and targeted methodologies and data analysis. It combines lectures and tutorial sessions to ensure a thorough understanding of the theory and practical applications. Topics covered include:
or contact Dr Liz Want (iptc@imperial.ac.uk) for further information. |
| 11-13 May 2017 |
Metabolism in Time and Space: Emerging Links to Cellular and Developmental Programs Venue: EMBL Heidelberg, Germany T. Alexandrov, A. Aulehla, P. Dorrestein, O. Leyser, S. McKnight, N. Perrimon Deadlines
Topics
Latest News
Stay up to date! Add this event
easily to your calendar by
downloading the
iCal>>
Add
to calendar For more information, visit http://www.embo-embl-symposia.org/symposia/2017/EES17-01/index.html
|
| 17-18 May 2017 |
Conference on Food and Nutritional Metabolomics for Health Venue: The Ohio State University, Columbus, OH, USA The purpose of this two-day event is to disseminate state-of-the-art knowledge in the field of food and nutritional metabolomics and foster networking and collaboration among colleagues and industry partners. For more information please visit: https://discovery.osu.edu/focus-areas/foods-for-health/events/conference-2017.html. |
| 29 May to 2 June 2017 |
W4E2017 Course: Analyze your LCMS, GCMS and NMR data with Galaxy and the Workflow4Metabolomics e-infrastructure Venue: Paris, France  The next Workflow4Experimenters international course (W4E2017) will take place in Paris (May 29 to June 2, 2017). During this one-week course (entirely in English), you will learn how to use Galaxy and the W4M infrastructure to analyze your own LC-MS, GC-MS, or NMR data set. Morning sessions will be dedicated to methodology and tools. Afternoon sessions will be devoted to tutoring. Invited speakers: Tim Ebbels (Imperial College), Steffen Neumann (IPB Halle), and Ralf Weber (Birmingham University). Registrations: http://workflow4metabolomics.org Contact: contact@workflow4metabolomics.org |
| 26-29
June
2017 |
Metabolomics 2017 Venue: Brisbane, Australia  It is our pleasure to invite you to the 13th International Conference of the Metabolomics Society from 25-29th June, 2017 at the Brisbane Conference and Exhibition Centre (BCEC) in Brisbane, Australia. Brisbane is a vibrant, friendly, lifestyle city—home to leading medical research and a thriving industry hub, located in the heart of Australia’s premier tourist region. The BCEC is rated among the top three convention centres in the world, and was the venue of the 2014 G20 Leaders Summit. It is ideally located in the unique riverside cultural and lifestyle precinct at South Bank, which is an inner city oasis with riverfront parkland, rainforest pockets and Australia’s only city-based sand and swimming beach as well as Australia’s newest and largest Gallery of Modern Art, cafes, restaurants and stylish shops. The conference has the theme of Building Bridges and under this banner extends its reach to the systems biology / genome-scale modelling community, as well as to the analytical chemistry / natural products chemistry community. In addition, the program features thematic streams for advancing the field, for food and environmental metabolomics, and for health and wellness. In addition, a deeper engagement between researchers within the Asia Pacific region is a natural focus for a conference held in Brisbane to promote metabolomics research, build and strengthen networks in the region. We invite you to attend an exciting scientific program comprising 27 oral sessions, 5 plenaries, 4 poster sessions, sponsored luncheons, as well as several keynote lectures and workshops. We will continue the successful tradition of satellite workshops to the conference in the afternoon of Sunday 25th June and the morning of Monday 26th June. Additionally, we have planned a range of social activities, including a welcome reception, an early-career researcher mixer and a conference dinner in the iconic BCEC Plaza Ballroom to give you a true Aussie-style experience. Brisbane is the ideal opportunity for delegates to enjoy a microcosm of Australia’s iconic experiences. World heritage listed rainforests, amazing beaches, islands, wineries and the internationally famous Australia Zoo—home of the crocodile hunter—are all easily accessible within an hour of the city. You can even do day trips to the Barrier Reef from Brisbane. On behalf of the Local Organising Committee and the Metabolomics Society Board we are excited to once again invite you to Metabolomics 2017—we are looking forward to welcoming you down under! Prof. Melissa Fitzgerald, School of Agriculture and Food Sciences & Dr. Horst Joachim Schirra, Centre for Advanced Imaging, The University of Queensland, Brisbane, Australia For more information, visit http://metabolomics2017.org/. |
| 11-13 Dec 2017 |
MetaboMeeting 2017 Venue: University of Birmingham, UK Make plans to attend the 10th successful MetaboMeeting conference. The meeting will bring together research scientists and practitioners from all areas of application and development of metabolic profiling, covering a wide range of experience from early career scientists to experts from throughout the international metabolomics field. MetaboMeeting 2017 continues to highlight the work of its attendees through both oral platform presentation and poster sessions. The deadline for oral presentation abstracts is 15th July 2017. The deadline for poster abstracts is 1st October 2017. For further information, visit http://metabomeeting2017.thempf.org/. |
Metabolomics Jobs |
This is a resource for
advertising positions in metabolomics. If
you have a job you would like posted in this
newsletter, please email Ian Forsythe (metabolomics.innovation@gmail.com).
Job postings will be carried for a maximum
of four issues (eight weeks) unless the
position is filled prior to that date.
Jobs Offered
| Job Title | Employer | Location | Posted | Closes | Source |
|---|---|---|---|---|---|
| Researcher Focusing on Metabolomics |
Uppsala University | Uppsala, Sweden | 31-Jan-2017 | 20-Feb-2017 |
Uppsala University |
| Research Associate in
Chemometrics |
Imperial College London | London, UK | 24-Jan-2017 | 23-Feb-2017 |
Imperial College London |
| Research Fellow in Mass
Spectrometry Metabolomics |
University of Birmingham | Birmingham, UK | 23-Jan-2017 | 23-Feb-2017 | University of Birmingham |
| Analytical Scientist for
Mass Spectrometry Based
Metabolomics/Lipidomics |
Biocrates Life Sciences AG | Innsbruck, Austria | 18-Jan-2017 | |
Biocrates Life Sciences AG
|
| Postdoctoral Fellowship
in Metabolomics |
Lund University Diabetes Centre | Malmö, Sweden | 21-Dec-2016 | |
Lund University Diabetes Centre |
|
Ian J. Forsythe Double AB, MSc Editor, MetaboNews Department
of Computing Science
University of Alberta 221 Athabasca Hall Edmonton, AB, T6G 2E8, Canada Email: metabolomics.innovation@gmail.com Website: http://www.metabonews.ca LinkedIn: https://ca.linkedin.com/in/ian-forsythe-465512128 Twitter: http://twitter.com/MetaboNews Google+: https://plus.google.com/118323357793551595134 Facebook: http://www.facebook.com/metabonews |
This newsletter is
published in partnership between The
Metabolomics Innovation Centre
(TMIC, http://www.metabolomicscentre.ca/) and
the Metabolomics Society (http://www.metabolomicssociety.org)
for the benefit of the worldwide
metabolomics community.
|