Understanding and Classifying Metabolite Space and
Metabolite-Likeness
Feature
article contributed by Julio E. Peironcely,
PhD student on Computational Metabolomics and Metabolite
Identification, at TNO,
Leiden University
and the Netherlands
Metabolomics Centre, The Netherlands
Original article: Peironcely
JE, Reijmers T, Coulier L, Bender A, Hankemeier T. Understanding
and classifying metabolite space and metabolite-likeness. PLoS
One. 2011;6(12):e28966. Epub 2011 Dec 14.
[
PMID:
22194963]
In metabolomics there is a recurrent bottleneck: metabolite
identification. Knowing the identity, i.e., the chemical
structure, of the detected compounds is essential to interpret the
experimental results.
Candidate structures that match experimental data are proposed
either by querying molecular databases or by using chemical
structure generators. In either case, the expert can face a large
list of compounds, from which to chose the correct structure. This
list could be ranked according to the metabolite-likeness of the
compounds.
With this work we wanted to answer two questions:
- What are the common characteristics of human metabolites?
- Can we predict the metabolite-likeness of a chemical
structure?
This work describes the application of computational tools to
predict the metabolite-likeness score of a molecule, i.e., how
much it resembles a metabolite, and to acquire a global
understanding of how the space of human metabolites is organized.
Datasets
Since we wanted to compare metabolites with non-metabolites, two
molecular datasets were selected. Human metabolites from the
HMDB were used as the metabolite set and commercially available
compounds listed in ZINC database were the non-metabolite dataset.
Molecules present in both datasets were removed, as well as
molecules in the HMDB that were described as drugs.
What are the common
characteristics of human metabolites?
Physicochemical properties
Several physicochemical properties were calculated for the HMDB
and ZINC datasets. Principal component analysis (PCA) was
performed. The result of this PCA (
Figure
1) reveals that both datasets cannot be completely separated
on the basis of physicochemical properties. This lack of
separation observed in the PCA, made us think that the utilization
of more sophisticated methods was required.
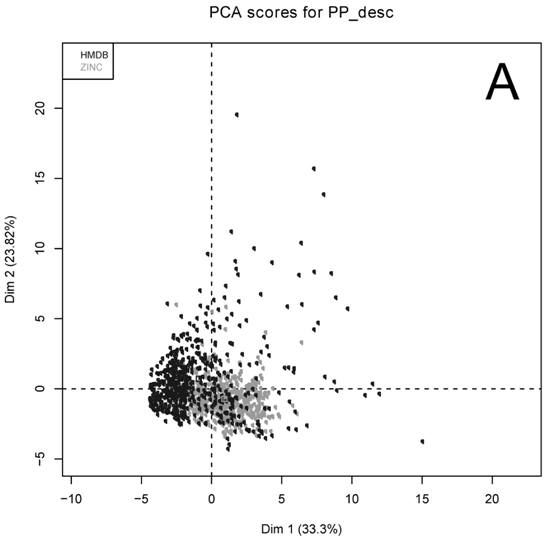
Figure 1. Principal
component analysis of the HMDB and ZINC datasets.
By looking at the properties that contributed the most to
the slight separation in the PCA (
Figure
2), we concluded that metabolites are more soluble in water,
have a lower molecular weight, have less complex structures, and
fewer carbon atoms than non-metabolites. Furthermore, Polar
Surface Area (PSA) tends to be larger in metabolites, suggesting
that they do not penetrate cell membranes as efficiently as
non-metabolites.

Figure 2. Physicochemical
properties contributing most to the variance in the first two
principal components of the PCA.
Fingerprint
Features and Frequent Fragments
Molecules from the HMDB and ZINC were described using ECFP_4
features, which can be seen as substructures, and partitioned
using a Classification Tree (
Figure
3). At each branching point of the tree, a feature is
chosen when it maximizes the split between metabolites and
non-metabolites.
Click on the thumbnail
below to view a larger version of the image
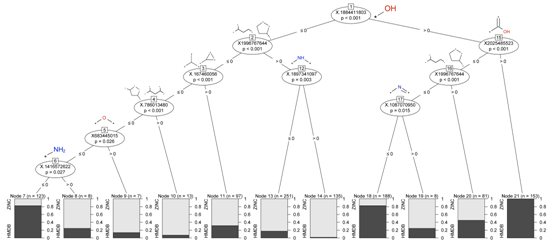
Figure 3. Classification
tree of the ECFP_4 features of the HMDB and ZINC datasets.
From this we learn that hydroxyl groups
are characteristic of metabolites. Also, nitrogen-containing
moieties correlate with non-metabolites. Acyclic molecules lacking
hydroxyls are likely to be metabolites.
Can we predict the
metabolite-likeness of a chemical structure?
Building the model
When classifying molecules
in
silico, different accuracies are obtained for each
representation and algorithm used. Thus, we selected five
different molecular representations (Atom Counts, Physicochemical
Properties, MDL Public Keys, ECFP_4 and FCFP_4 fingerprints) and
three different classification algorithms (Support Vector Machines
(SVM), Random Forest (RF), Naïve Bayes Classifier (NB)). Our
aim was to employ as our model of Metabolite-Likeness the
combination reporting the highest accuracy in the predictions.
The process of model building is depicted in
Figure
4. A standardization process cleans the molecules from both
databases. Diversity selection is performed to reduce the impact
that overrepresented families of metabolites, for instance lipids,
might have in the model. This involved clustering each dataset,
where the cluster centers were collected as the training set and
the other compounds were used as the test set.
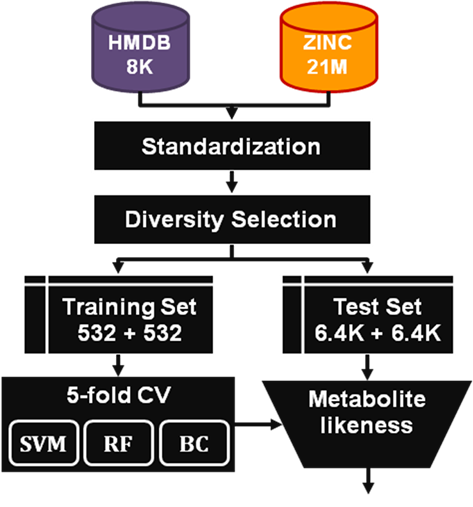
Figure 4. Model
building workflow.
Since SVM and RF classifiers require the tuning of some
metaparameters, we performed a five-fold cross validation to
select their optimal values. Once we had the metaparameters,
fifteen models (five representations x three classifiers) were
built. These models were used to classify the molecules in the
test set and, as a result, the best performing model was the one
built with Random Forest and MDL Public Keys. It classified
correctly 99.84% of the metabolites and 88.79% of the
non-metabolites.
Prospective validation of the
model
A common problem of predictive models is how they perform when
predicting new objects. Hence we compiled three prospective
validation sets to test the quality of our best model. The first
dataset contained 457 metabolites that had not yet been included
in the HMDB. The second dataset represented non-metabolites and
included molecules from ChEMBL, which can be seen as background
dataset alternative to ZINC. The third dataset were drugs from
DrugBank. These were selected to test the hypothesis that many
drugs tend to mimic metabolites, or at least they are more
metabolite-like than screening compounds.
The results (
Figure
5) showed that 95.84% of the new metabolites were predicted
correctly. Concerning drugs, 54.3% obtained a metabolite-likeness
of 50% or higher, which was in accordance with our assumption that
many drugs resemble metabolites. For the ChEMBL compounds, only
22.29% were predicted as metabolites.
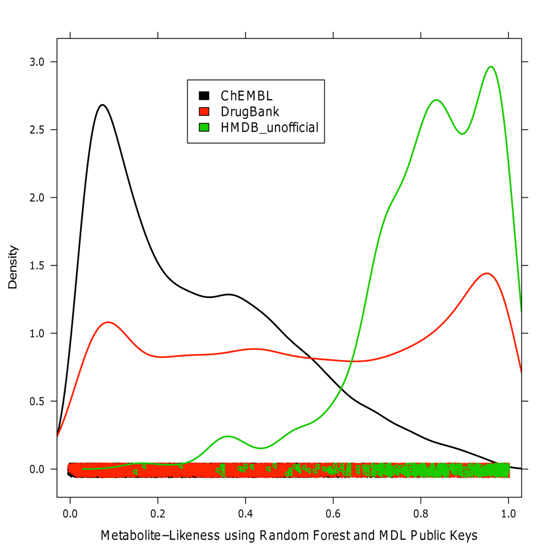
Figure 5. Metabolite-likeness
distribution of the prospective validation sets.
Molecules of the three different classes,
which fall into different bins of metabolite-likeness scores, are
presented in
Figure
6. The first noticeable feature is the absence of a
metabolite with a predicted metabolite-likeness smaller than 10%,
underlining the homogeneity of metabolites as a class. Metabolite
HMDB13193 obtained the lowest metabolite-likeness, 17%, and
contains two chlorine atoms, which is not common in metabolites.
Another interesting situation occurs with molecules that have a
steroid scaffold, a common fragment in endogenous metabolites.
Drug DB00180 (flunisolide) obtains a metabolite-likeness value of
52%. It possesses a fluorine atom, which is not frequent in
metabolites, and which might have reduced its metabolite-likeness
score. Conversely, ChEMBL compound CHEMBL1163241 also has the
steroid scaffold but obtains a score of just 35.2% on the
metabolite-likeness scale, due to having two fluorine atoms and a
secondary amine, features that the classification tree revealed to
be common in non-metabolites. Compounds with high-predicted
metabolite-likeness are DB00131 (adenosine monophosphate), DB00125
(L-arginine), CHEMBL6422, and CHEMBL14568, which receive 84.2%,
99%, 82.8%, and 96.8%, respectively. Adenosine monophosphate
includes the phosphate group, frequently found in metabolites
together with two hydroxyl groups. Metabolite-like features of
L-arginine, like linearity and a carboxylic group, outweigh the
non-metabolite features like the nitrogen-containing functional
groups. While compound CHEMBL6422 possesses carboxylic acid and
hydroxyl functionalities, CHEMBL14568 is small, linear, and also
exhibits a hydroxyl group, leading to a very high
metabolite-likeness score.
Click
on the thumbnail below to view a larger version of the image
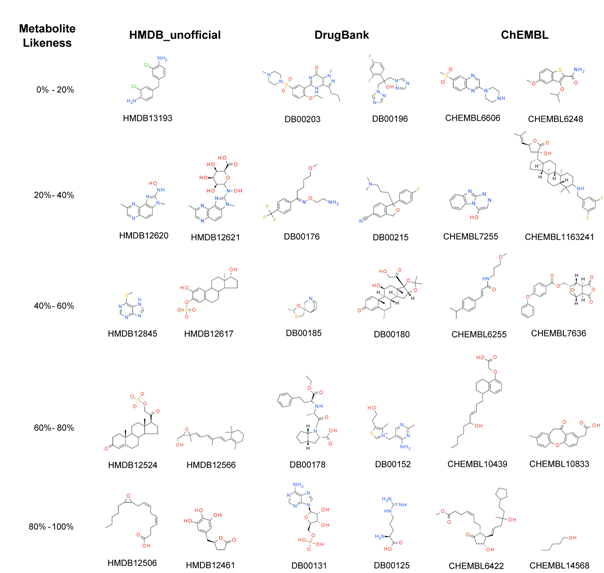
Figure 6. Molecules
of the prospective validation sets with different predicted
metabolite-likeness values.
These results demonstrate that our model is successful at
identifying whether a molecule is a metabolite or not. We expect
metabolite-likeness prediction to help studies that involve
metabolite identification in the future, when no database match is
found for the unknown compound. Then the candidate structures are
generated based on mass spectrometry data, e.g., elemental
composition, using a structure generation tool. These output
molecules would be ranked according to their metabolite-likeness.
Please
note: If you know of any
metabolomics research programs, software, databases,
statistical methods, meetings, workshops, or training
sessions that we should feature in future issues of this
newsletter, please email Ian Forsythe at metabolomics.innovation@gmail.com.
Feature article contributed by Ian
Forsythe, Editor, MetaboNews, Dept of Computing
Science, University of Alberta, Edmonton, Canada
Metabolomics
is an emerging field that is complementary to other omics
sciences and that is gaining increasing interest across all
disciplines. Because of metabolomics' unique advantages, it is
now being applied in functional genomics, integrative and
systems biology, pharmacogenomics, and biomarker discovery for
drug development and therapy monitoring. More than 95% of
today's biomarkers are small molecules or metabolites (MW
<1500 Da), which can be used for disease testing, drug
testing, toxic exposure testing, and food consumption
tracking. While standard clinical assays are limited in the
number and type of compounds that can be detected,
metabolomics measures many more compounds. Since a single
compound is not always the best biomarker (diagnostic,
prognostic or predictive), healthcare practitioners can use
metabolomics
information about multiple compounds to make better medical decisions.
Global metabolic profiling is now being used to determine
clinical biomarkers in assessing the pathophysiological health
status of patients.
In the following two recent studies, metabolomics approaches
were used to develop biomarker tools for the identification of
biomarkers associated with diabetic kidney disease and ovarian
cancer, respectively.
1. van der Kloet FM, Tempels FWA,
Ismail N, van der Heijden R, Kasper PT, Rojas-Cherto M, van
Doorn R, Spijksma G, Koek M, van der Greef J, Mäkinen VP,
Forsblom C, Holthöfer H, Groop PH, Reijmers TH, Hankemeier
T. Discovery of early-stage biomarkers for diabetic kidney
disease using ms-based metabolomics (FinnDiane study).
Metabolomics. 2012 March, Volume 8, Number 1, 109-119, DOI:
10.1007/s11306-011-0291-6 [
Metabolomics]
In
this paper, the research team performed metabolite profiling
to identify early-stage biomarkers for diabetic kidney disease
(DKD) using gas chromatography-mass spectrometry (GC-MS) and
liquid chromatography-mass spectrometry (LC-MS). A clinical
indicator of DKD is abnormally high urinary albumin excretion
rates. The goal of the study was to uncover urinary biomarkers
that allow one to differentiate between the progressive and
non-progressive forms of albuminuria in humans. As a result of
this work, the researchers pinpointed acyl-carnitines,
acyl-glycines, and metabolites related to tryptophan
metabolism as the differentiating biomarkers.
2. Fong MY, McDunn J, Kakar SS. Identification of
metabolites in the normal ovary and their transformation in
primary and metastatic ovarian cancer. PLoS One.
2011;6(5):e19963. Epub 2011 May 19. [
PMID:
21625518]
This research team sought to characterize the human ovarian
metabolome and identify disease-associated changes due to
primary epithelial ovarian cancer (EOC) and metastatic
ovarian cancer (MOC). The team used three analytical
platforms: 1. gas chromatography mass spectrometry (GC/MS),
2. liquid chromatography tandem mass spectrometry (LC/MS/MS)
set up to catalog positive ions, and 3. LC/MS/MS set up to
catalog negative ions. According to this paper, the human
ovarian metabolome consists of 364 biochemicals. Upon
transformation of the ovary, two energy utilization pathways
were perturbed, namely β-oxidation of fatty acids and
glycolysis, and the following metabolites were present at
increased levels in EOC and MOC: carnitine, acetylcarnitine,
and butyrylcarnitine. In EOC, there were significant
increases in phenylpyruvate and phenyllactate. In both EOC
and MOC, the research team also found increased levels of
2-aminobutyrate. Metabolites found at increased levels in
various disease states may serve as candidate biomarkers for
the progression of ovarian cancer.

|
3) Metabolomics Current Contents
|
Recently published papers in metabolomics:
 |
4)
MetaboNews
|
30 Jan 2012
|
Genetic regulation of
metabolomic biomarkers – paths to cardiovascular
diseases and type 2 diabetes - The research group
at the Institute for Molecular Medicine Finland
(FIMM) has revealed eleven new genetic regions
associated with the blood levels of the
metabolites, including new loci affecting
well-established risk markers for cardiovascular
disease and potential biomarkers for type 2
diabetes. The findings may help in elucidating the
processes leading to common diseases. The study
will be published in Nature Genetics.
In a study to the genetic variance of human
metabolism, researchers have identified thirty one
regions of the genome that were associated with
levels of circulating metabolites, i.e., small
molecules that take part in various chemical
reactions of human body. Many of the studied
metabolites are biomarkers for cardiovascular
disease or related disorders, thus the loci
uncovered may provide valuable insight into the
biological processes leading to common diseases.
Laboratory tests used in the clinic typically
monitor one or few circulating metabolites. The
researchers at the Institute for Molecular
Medicine Finland (FIMM) used a high throughput
method called nuclear magnetic resonance (NMR)
that can measure more than hundred different
metabolites in one assay. This provides a much
more in-depth picture of circulating metabolic
compounds.
Paper: Kettunen J,
Tukiainen T, Sarin A-P, Ortega-Alonso A, Tikkanen E,
Lyytikäinen L-P, Kangas AJ, Soininen P,
Würtz P, Silander K, Dick DM, Rose RJ, Savolainen
MJ, Viikari J, Kähönen M, Lehtimäki T,
Pietiläinen KH, Inouye M, McCarthy MI, Jula A,
Eriksson J, Raitakari OT, Salomaa V, Kaprio J,
Järvelin M-R, Peltonen L, Perola M, Freimer NB,
Ala-Korpela M, Palotie A, Ripatti S. Genome-wide
association study identifies multiple loci influencing
human serum metabolite levels. Nature Genetics,
29th January, 2012 ( online).
|
27 Jan 2012
|
Giant Cell Reveals
Metabolic Secrets - Chemical reactions
within the cell produce intermediate and end
products in the form of small molecules called
metabolites. Using an approach called
metabolomics, a research team led by Kazuki Saito
of the RIKEN Plant Science Center, Yokohama and
Tsuruoka, has elucidated the localization and
dynamics of 125 metabolites within a single giant
cell of the freshwater alga Chara australis.
The team's findings provide important insights
into the fundamental processes of cells in
general.
Metabolites play important roles in the regulation
of critical biological processes, such as growth,
differentiation, and, in the case of so-called
'secondary metabolites', chemical defense.
"Metabolomics is the systematic study of these
unique chemical footprints, and involves
identifying and characterizing the many
metabolites found in a cell, tissue, organ or
organism, as well as their production,
distribution and dynamics," explains Saito.
Because the enzymes involved in producing and
converting different metabolites are often
localized within subcellular structures called
organelles, biologists generally assumed that
metabolites are also highly compartmentalized
within the cell, but none had demonstrated this
comprehensively.
"Understanding the compartmentalization and
dynamics of metabolites within single organelles
represents an enormous technical challenge, not
least because of the tiny size of these structures
in most cells," says Saito.
Paper:
Oikawa A, Matsuda F, Kikuyama M, Mimura T, Saito
K. Metabolomics of a Single Vacuole Reveals
Metabolic Dynamism in an Alga Chara australis.
Plant Physiology,
2011; 157 (2): 544 DOI: 10.1104/pp.111.183772
|
24 Jan 2012
|
Upregulated Metabolite May
Be Target for Neuropathic Pain Therapy -
Blocking
overproduction of the metabolite N,N-dimethylsphingosine
(DMS) in the spinal cord could feasibly represent a
therapeutic option for treating neuropathic pain,
researchers claim. A team at Washington University
School of Medicine, St. Louis, and The Scripps
Research Institute used an MS-based metabolomics
approaches to compare metabolite levels in specific
tissues of tibial nerve-transacted (TNT) experimental
rats and sham-operated control rats. Tissues analyzed
included spinal cord dorsal horn and dorsal root
ganglia, damaged tibial nerve, and blood plasma.
Their results, reported in Nature Chemical Biology by Gary
Siuzdak, Ph.D., and colleagues, demonstrated that 94%
of metabolite differences between TNT rats and control
rats occurred in the ipsilateral dorsal horn, rather
than in the damaged nerve itself. When the team
further characterized dysregulated metabolites in the
dorsal horn tissue, they identified multiple
alterations in sphingomyelin-ceramide metabolism 21
days after TNT injury. “We therefore hypothesized that
dysregulated metabolites in this pathway may be linked
to the physiological changes underlying neuropathic
pain and represent possible new targets for
therapeutic intervention,” they state.
The changes in dorsal horn metabolite concentrations
were consistent with degradation of sphingomyelin, and
included significant upregulation of DMS, which hasn’t
previously been investigated in the context of
neuropathic pain, the authors add. Of significant
interest was the finding that intrathecal injections
of DMS in healthy rats led to the animals developing
mechanical allodynia (pain resulting from a stimulus
that wouldn’t normally be painful) in the hind paw.
This effect occurred at injected concentrations of DMS
that resulted in similar dorsal horn levels of the
metabolite that were found in the TNT experimental
mice.
Paper: Patti
GJ, Yanes O, Shriver LP, Courade JP, Tautenhahn R,
Manchester M, Siuzdak G. Metabolomics implicates
altered sphingolipids in chronic pain of neuropathic
origin. Nat Chem
Biol. 2012 Jan 22. doi: 10.1038/nchembio.767.
[Epub ahead of print] [ PMID:
22267119]
Source: Genetic
Engineering & Biotechnology News
|
Please note:
If you know of any metabolomics news that we should feature
in future issues of this newsletter, please email Ian
Forsythe (metabolomics.innovation@gmail.com).
 |
5)
Metabolomics Events
|
| 3 Feb 2012 |
Cancer Metabolomics: Elucidating the
Biochemical Programs that Support Cancer Initiation
and Progression
Venue: New York, New York, USA
Since the 1920s, it has been recognized that cancer
cells exhibit metabolic features that are distinct from
those of 'normal' cells. However, a comprehensive
picture of cancer metabolism and its molecular
underpinnings has been essentially unapproachable—that
is, until very recently. With the emergence of effective
analytical strategies for broad-based metabolite
profiling (both targeted and untargeted), the cancer
cell 'metabolome' has now come into sight. Taking
advantage of LC-MS-based analytical platforms, the
participating speakers will describe new knowledge of
metabolic pathways that distinguish cancer cells,
signaling cascades that drive cancer-selective metabolic
pathways and implications for the development of novel
cancer chemotherapies.
Call for Poster
Abstracts
A poster session will be held and a selected number of
presenters will be asked to give brief oral
presentations. The deadline for abstract submission is
Friday, January 27, 2012. For abstract instructions,
send an email to CancerMetabolomics@nyas.org
with the words "Abstract Information" in the subject
line. Instructions will be forwarded automatically. For
questions, please call 212.298.8618.
|
| 20-22 Feb 2012 |
International Conference and Exhibition on
Metabolomics & Systems Biology
Venue: San Francisco, USA
OMICS Group invites you to attend the International
Conference and Exhibition on Metabolomics & Systems
Biology which is going to be held during 20-22 February
2012 San Francisco, USA.
Metabolomics-2012
will serve as a catalyst for the advances in the study
of Metabolomics & Systems Biology by connecting
scientists within and across disciplines at sessions and
exhibition held at the venue, creates an environment
conducive to information exchange, generation of new
ideas, and acceleration of applications that benefit
Research in Metabolomics & Systems Biology.
Conference Highlights the following topics:
- Proteomics & Genomics
- Transcriptomics & Metabolomics
- Bioinformatics
- Gene expression Profiling
- Immunology
- Microbiology & Biochemistry
- Computational Biology
- Genetics and Metabolism
- Glycomics & Lipidomics
|
| 21-22 Feb 2012 |
7th Annual Biomarkers Conference
Venue: Manchester, UK
The pharmaceutical industry is facing high clinical
development costs and declining drug discovery success
rates. To stay competitive companies are re-evaluating
their drug development process to reduce attrition
rates. Biomarkers promise to transform drug discovery,
clinical development and diagnostics in the R&D
process as effective use of biomarkers at each stage of
R&D can improve decision-making, increase clinical
trial success rates and productivity.
The 7th Annual
Biomarkers Congress 2012 presented by Oxford
Global Conferences provides a first class educational
and networking opportunity for over 250 attendees to
gain knowledge and insights into the Biomarkers
marketplace.
Do not miss out on the opportunity to learn how you can
streamline your R&D process and identify potential
cost savings through successful biomarker discovery,
validation and clinical development.
Our CNS
Biomarkers Congress 2012 which will be co-located
with the 7th Annual
Biomarkers Congress on the 21- 22 February will
provide the opportunity for two days worth of deep
analysis and insight into this growing CNS Biomarkers
market.
For more information, visit http://www.biomarkers-congress.com/.
|
| 12-14 Mar 2012 |
Metabolic Leaders' Forum 2012
Venue: San Francisco, California
Phacilitate’s inaugural Metabolic Leaders’ Forum
takes a timely look at the decisions facing senior
R&D executives following the tightening up of
requirements for approving new drugs for adult-onset
diabetes coupled with the FDA’s more recent rejection
of three separate drugs to treat obesity. What are
your peers doing to establish appropriate CV benefits
profiles to demonstrate positive outcomes without
incurring exploding costs? How are companies large and
small rethinking and restructuring to optimize their
ability to compete in diabetes and obesity, and the
emerging cardiometabolic health field?
For more information, visit http://www.phacilitate.co.uk/event.php?eid=9&pid=259&opstatus=confrence.
|
| 17-20 Apr 2012 |
analytica Conference 2012
Venue: Munich, Germany
For the classical exhibition area, the analytica
Conference provides the perfect complement. It has been
a decisive factor in establishing analytica as the
pre-eminent meeting point for the industry.
In various symposiums, leading scientists from all over
the world report on the latest developments, current
trends and visions of the future. Analytic, diagnostic,
biochemical and molecular biological methods and
procedures are discussed here. On the last occasion, 140
well-known experts gave talks in 23 different thematic
symposiums.
Main subject emphases/highlights of the analytica
Conference 2010
- Presentation of the Gerstel Award and the
Bunsen-Kirchhoff Award
- Patient-oriented laboratory diagnostics
- Separation techniques in the life sciences
- Doping analytics
- Proteome research
- Measurement and toxicology of particulate matter
- Modern analytical methods for the chemical
analysis of art objects
- Analytical contributions to the treatment of
diabetes
The 2012 event will focus on topics such as acute
diagnostics and clinical metabolomics.
|
| 7-8 May 2012 |
LIPID MAPS Annual Meeting 2012: Lipidomics
Impact on Cell Biology, Metabolomics and
Translational Medicine
Venue: La Jolla, CA, USA
This is an exciting time for the emerging field of
lipidomics. With the development and evolution of
sophisticated mass spectrometers linked to highly
efficient liquid chromatography systems, individual
molecular species of lipids can now be isolated and
identified, allowing us to begin to understand lipid
metabolism and the treatment of lipid-based diseases
(atherosclerosis and inflammatory disease as well as
arthritis, cancer, diabetes and Alzheimer's disease).
Recent awareness that each category of lipid consists of
thousands if not tens of thousands of individual
molecular species requires sophisticated informatics to
ensure consistent databasing and annotation of the
numerous lipid molecular species and analysis of
tremendous quantities of experimental data. The goal of
this meeting is to bring together biological and
biomedical scientists in a wide range of fields to share
new findings and methods in the broader lipidomics field
and to explore joint efforts to extend the use of these
powerful new methods to new applications. Presentations
will provide an excellent introduction for scientists
new to these methods, and are sure to be of interest to
lipidomics veterans to learn about latest techniques and
research results.
The meeting program tentatively features the following
six sessions:
- Cancer
- Inflammation
- Macrophage Biology
- Metabolic Disease
- Metabolomics
- Translational Medicine
|
| 20-24 May 2012 |
60th ASMS Conference on Mass Spectrometry and
Allied Topics
Venue: Vancouver, BC, Canada
The conference and short courses will be held at the
Vancouver Convention Centre, 1055 Canada Place,
Vancouver, BC V6C 0C3, Canada. All oral sessions,
poster sessions, exhibit booths, and corporate
hospitality suites will be located in the Convention
Centre.
Important Dates:
Jan 9, 2012:
Conference registration and lodging opens online
Feb 3, 2012: Deadline for submission of
abstracts
Apr 14, 2012: Conference program online
Apr 30, 2012: Deadline for advance registration
|
| 21-23 May 2012 |
Les 6èmes Journées
Scientifiques du Réseau Français de
Métabolomique et Fluxomique (JS 6 RFMF)
Venue: Nantes, France
The 6th Scientific Days of the French Network of
Metabolomics and Fluxome will be held in Nantes from
21 May 2012 to May 23, 2012, organized by the platform
Corsaire, the platform of Metabolomics Biogenouest.
Corsair includes eight technical support centers
located throughout Western France (Brest, Nantes,
Rennes, and Roscoff).
The main topics selected for JS 6 RFMF are:
- Applications of metabolomics and fluxomics in the
areas of the sea, agronomy and health.
- Technological developments, bioinformatics, and
statistical processing
For more information, please visit https://colloque4.inra.fr/6_js_reseau_francais_metabolomique_fluxomique.
|
| 25-28 June 2012 |
METABOLOMICS 2012: Breakthroughs in plant,
microbial and human biology, clinical and
nutritional research, and biomarker discovery
Venue: Washington Marriott Wardman Park Hotel,
Washington, DC, USA
The annual meeting of the Metabolomics Society brings
together metabolomics researchers from around the world
to discuss their most recent achievements as they work
to harness the power of metabolomics. Gathering together
in a hospitable venue is key to developing the collegial
interactions that can build a successful community that
advances together.
The 2012 meeting promises a program full of practical
workshops and parallel sessions covering the broad range
of biological and technological metabolomics topics as
well as providing rich opportunities for networking.
Join us as we gather together to share ideas, insights,
advances and obstacles in the multi-faceted world of
metabolomics.
|
| 10 July 2012 |
Merck Deep Dive: Translational Technologies
for Basic and Clinical Scientists
Venue: Ocean Place Resort and Spa, Long Branch, New
Jersey, USA
Join us for another in this quarterly unique series that
brings together about 150 Merck subject experts and key
vendors. Sponsors and Exhibitors can display products
and services to a very focused set of the Merck
community leaders. Select exhibitors are given an
opportunity to privately present upcoming product
development and get instant feedback from Merck on where
they would like to see development go to meet their
upcoming needs. Topic for this event: Translational
Technologies for Basic and Clinical Scientists.
Molecular BioMarkers (Proteomics/Genomics/Metabolomics)
is a key part of this event.
|
Please
note: If you know of any metabolomics lectures,
meetings, workshops, or training sessions that we should
feature in future issues of this newsletter, please email
Ian Forsythe (metabolomics.innovation@gmail.com).
 |
6)
Metabolomics Jobs
|
This is a resource for advertising
positions in metabolomics. If you have a job you would like
posted in this newsletter, please email Ian Forsythe (metabolomics.innovation@gmail.com).
Job postings will be carried for a maximum of 4
issues (8 weeks) unless the position is filled prior to that
date.
Jobs
Offered
| Job Title |
Employer |
Location |
Date Posted |
Source |
Senior Principal
Scientist, PepsiCo R&D Long Term Research Biology
Innovation Lab
|
PepsiCo
|
700 Anderson Hill Road Purchase, New York
10577 |
27-Jan-2012
|
Metabolomics
Society Jobs |
Senior Specialist
in Metabolomics instrumental analysis
|
IMDEA Food Institute
|
Madrid, Spain |
16-Jan-2012 |
European
Commission Jobs
|
| Postdoctoral
Research Fellow in the Sumner lab focused on
Metabolomics to Elucidate Genes of Unknown Function
funded by NSF 2010 (PB-S095-570) |
The Samuel Roberts Noble Foundation |
Ardmore, OK 73401, USA |
5-Jan-2012 |
Metabolomics
Society Jobs |
| Postdoctoral
Research Fellow in the Sumner lab focused on
Computational Annotation of the Medicago and
Arabidopsis Plant Metabolomes funded by a
collaborative award from NSF & JST (PB-S095-700) |
The Samuel Roberts Noble Foundation |
Ardmore, OK 73401, USA |
5-Jan-2012 |
Metabolomics
Society Jobs |
| Postdoctoral
Research Fellow in the Sumner lab focused on Empirical
Annotation of the Medicago and Arabidopsis Plant
Metabolomes funded by a collaborative award from NSF
& JST (PB-S095-701) |
The Samuel Roberts Noble Foundation |
Ardmore, OK 73401, USA |
5-Jan-2012 |
Metabolomics
Society Jobs |
| Research
Associate in the Analytical Chemistry Core Facility
focused on GCMS Metabolomics Applications
(PB-S070-125) |
The Samuel Roberts Noble Foundation |
Ardmore, OK 73401, USA |
5-Jan-2012 |
Metabolomics
Society Jobs |
| NMR
Instrument Coordinator in the Sumner lab focused upon
the successful installation and integration of a new
NMR with UHPLC-MS and automated solid phase extraction
(i.e., UHPLC-MS-SPE-NMR) |
The Samuel Roberts Noble Foundation |
Ardmore, OK 73401, USA |
5-Jan-2012 |
Metabolomics
Society Jobs |
| Postdoctoral
Researcher in Molecular (Omics) Nanotoxicology |
University of Birmingham |
Birmingham, UK |
4-Jan-2012 |
Metabolomics
Society Jobs |
| Metabolomics
Tenured Faculty Position |
UNC Chapel Hill |
Kannapolis, North Carolina |
4-Jan-2012 |
Scientific
American Jobs |
| Postdoctoral
researcher in environmental metabolomics |
University of Birmingham |
Birmingham, UK |
15-Dec-2011
|
Metabolomics
Society Jobs |
| Post-doctoral
position (18 months) - Cancer, Environment and
Metabolomics |
LABERCA
|
Nantes, France
|
13-Dec-2011
|
Réseau
Français de Métabolomique et Fluxomique |
Jobs Wanted
This
section is intended for very highly qualified individuals
(e.g., lab managers, professors, directors, executives with
extensive experience) who are seeking employment in
metabolomics. We encourage these individuals to submit their
position requests to Ian Forsythe (metabolomics.innovation@gmail.com).
Upon
review, a limited number of job submissions will be selected
for publication in the Jobs Wanted section.
Note: There are no postings at
this time.
Free
Subscription
















