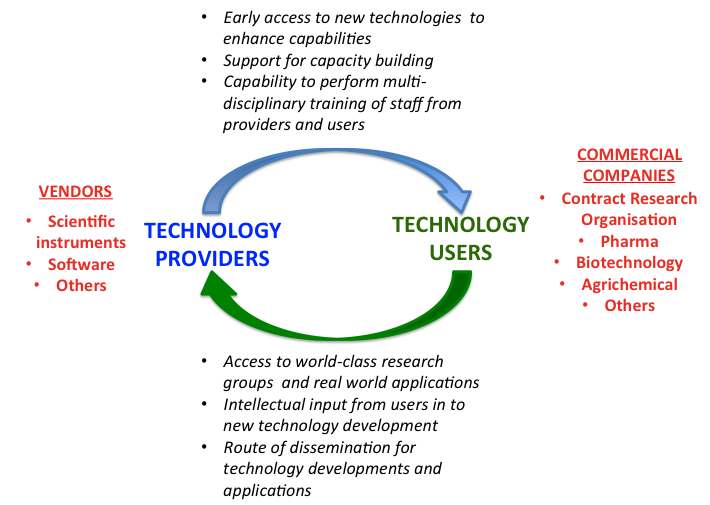
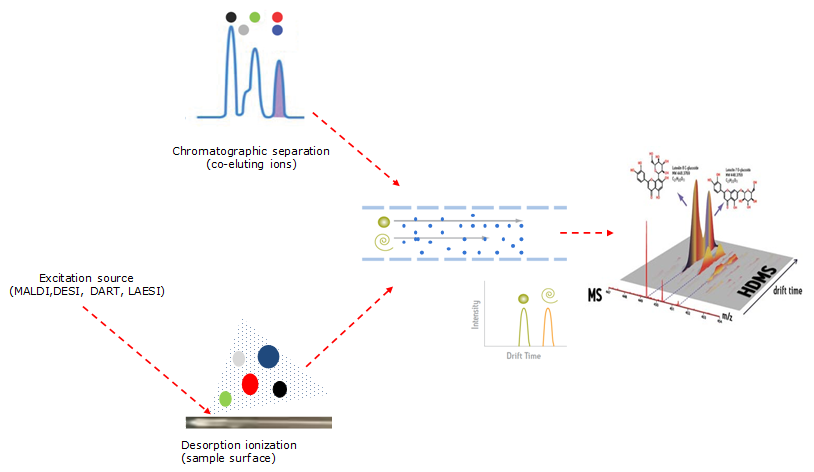
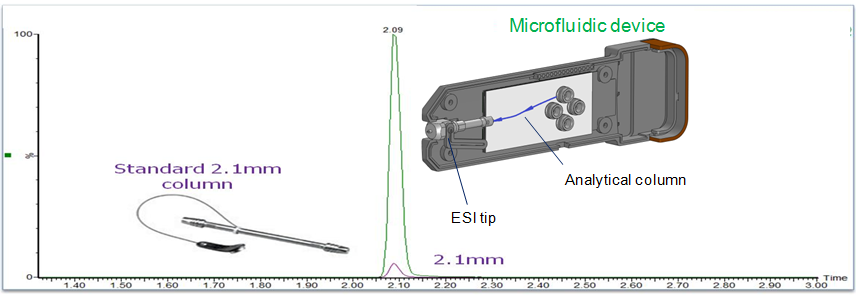
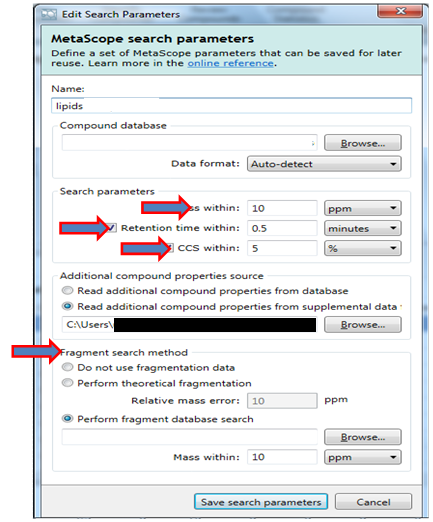
Enhancing Colon
Cancer Screening through Polyp Detection
Metabolomic
Technologies (MTI) has developed a
technology that detects colonic polyps, the
precursor to colorectal cancer (CRC), using a
noninvasive urine test. Unlike fecal-based
tests, PolypDx™ detects polyps before they
become cancerous, according to the company. The
assay, now in beta testing, has the potential to
improve patient compliance. By enabling early
intervention, it also may reduce the high
healthcare costs associated with treating CRC,
by reducing the numbers of surgeries and
hospitalizations.
“In Canada, colonoscopies are not the first line
of defense. Instead, our healthcare system uses
fecal-based tests,” Reg Joseph, CEO, explains.
For those tests, patients collect their samples
at home and send them to a lab. With that
approach, compliance rates across Canada are as
low as 14%. “Because the fecal tests detect CRC
only 50% of the time, often people without
polyps and cancer undergo colonoscopies. Eighty
percent of patients referred for colonoscopies
that are based upon fecal tests do not have CRC.
Because of the invasive nature of this procedure
and lack of patient compliance, many are not
screened in time.”
PolypDx addresses those issues, MTI reports. It
is a single spot test that analyzes metabolites
in the urine to determine whether patients have
CRC and, more importantly, whether they have
colonic polyps. The test has a sensitivity of
71% and a specificity of 61% in terms of polyp
detection. In contrast, standard-of-care
fecal-based tests are designed to detect CRC—not
colonic polyps. When used to detect polyps,
fecal-based tests have sensitivities of 1–15%.
Therefore, PolypDx represents a significant
advance in polyp detection.
“We’re continuing to improve sensitivity and
specificity for PolypDx by screening against a
larger metabolomic library,” Joseph says. The
current version is based on an NMR platform. An
improved version offering higher throughput and
enhanced ease of use is being developed on a
mass spectroscopy platform. It is expected to
become available in 2015. Mass spec instruments
are used routinely in large diagnostics labs, so
this new test kit can be integrated easily into
current workflows.
“We’re working with Alberta Health Services and
DynaLIFEDx to look at scaling up this test for
large population screening,” Joseph says.
Alberta Health Services is collaborating with
MTI to release the assay kit as early as
possible. MTI plans to commercialize the first
version of the test in late 2014.
Ultimately, MTI envisions PolypDx as a routine
screening tool that will be used before patients
undergo a colonoscopy. “We’d like to be able to
screen thousands of patients per day using one
instrument,” Joseph says.
“More than 95% of colon cancer patients develop
cancer through adenomatous colonic polyps. If we
can identify these patients before the onset of
cancer, the chance of survival is more than
90%.” The PolypDx assay kit is expected to be
priced at about $50. The annual U.S. burden for
CRC treatment is $14 billion.
MTI’s platform also may be harnessed for other
diseases. MTI is developing ColoDx™ for CRC.
Additionally, “We can see the preliminary
signatures for prostate and breast cancers.”
Urine-based assays for those diseases are in the
early development phase.
The technology spun out of the University of
Alberta in Canada, which is known globally for
metabolomics research. Company founders,
gastroenterologist Richard Fedorak, M.D., and
colorectal surgeon Haili Wang, M.D.,
collaborated with the university’s metabolomics
group, which was collecting bio-samples to
identify potential diagnostics for a variety of
diseases. A retrospective study led to a
prospective study and, eventually, PolypDx.
“MTI is actively seeking partnerships and
strategic commercial partners,” Joseph says.
MTI is collaborating with BGI-Shenzhen to
develop the assays for the Chinese market. The
assay is undergoing validation and clinical
trials in China. According to Yong Zhang, Ph.D.,
head, proteomic division, BGI-Shenzhen, “Our
company is the best positioned to co-develop
MTI’s diagnostic tests for the Chinese market,
assist with the regulatory process, and market
the technology.”
“In the West, the regulatory landscape for
multivariate tests like metabolomics is a bit
gray. China, however, has a progressive
regulatory framework that is focused around
driving down the costs of disease for a large,
new middle class that is demanding high-quality
healthcare.” Therefore, “Preventive
strategies—including diagnostics—are on the fast
track.” Additionally, the University of Alberta
has had research collaboration with BGI-Shenzhen
for the past five years. Leveraging that
relationship enables MTI to showcase a
translational project and make a real
difference, quickly, in individuals’ health.
Joseph says MTI’s finances are sound. MTI is
funded by angel and super angel investors and by
grants from Alberta Innovates—Health Solutions,
the National Research Council of Canada’s
Industrial Research Assistance Program, and
Alberta Enterprise and Advanced Education. “Our
burn rate isn’t excessively high, so regional
financing will take us through the next few
years.”