
TMIC and the Metabolomics Society
Issue 41 - January 2015
CONTENTS:
Online version of this newsletter:
http://www.metabonews.ca/Jan2015/MetaboNews_Jan2015.htm
 |
| Published
in partnership between TMIC and the Metabolomics Society Issue 41 - January 2015 |
|
CONTENTS: |
|
|
 |
Metabolomics Society News |
3rd
International Conference on Analytical
Science & Technology Dr.
Geum-Sook Hwang, organizer of the conference, KBSI Dr.
Geum-Sook Hwang, organizer of the conference, KBSI |

| |
Method Spotlight
|
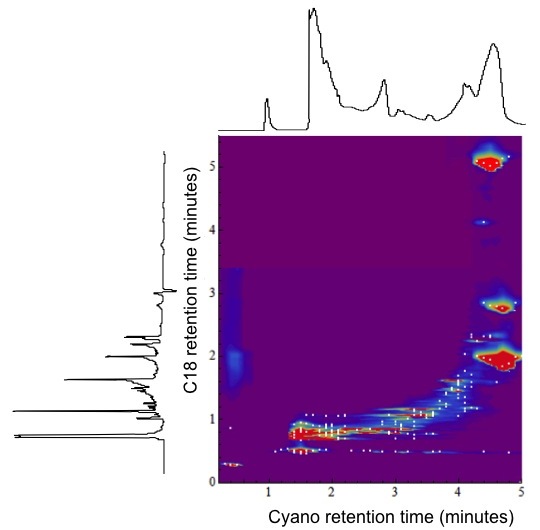
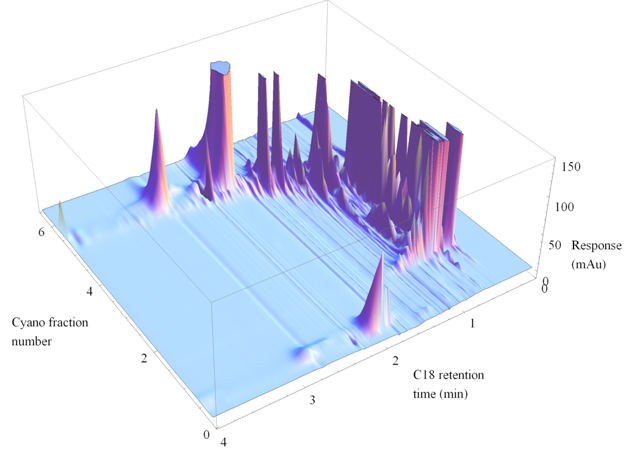

| |
MetaboInterviews
|
| Professor, Department of Agricultural, Food & Nutritional Science, University of Alberta, Edmonton, Canada |
|
 |
Biography Dr. Ametaj received his doctorate degree in nutritional immunology from Iowa State University and did three postdoctoral trainings at Iowa State University, Purdue University, and Cornell University before joining the University of Alberta in 2004. Dr. Ametaj’s research interest is in the area of nutritional immunology. His long-term goals are to study the relationship between nutrition and immune responses, and their contribution in development of production diseases in ruminant animals and to develop new strategies to reduce the high incidence of periparturient diseases in dairy cattle. Dr. Ametaj’s research areas include: 1) determining dairy cattle metabolome in four major fluids (milk, plasma, urine, and rumen fluid), 2) identifying potential biomarkers of disease in major fluid of transition dairy cows, 3) developing a new vaccine for prevention of transition cow diseases, 4) using probiotics to prevent uterine infections and improve reproductive and productive performance of dairy cows, and 5) studying the etiology and pathogenesis of transmissible spongiform encephalopathies. |
 |
Metabolomics
Current Contents
|
 |
MetaboNews |
| 31
Dec 2014 |
LECO's ChromaTOF-HRT®
1.81 Now Available
LECO Corporation is pleased to announce the availability of a major upgrade to its ChromaTOF-HRT brand software platform. ChromaTOF-HRT version 1.81 offers both new features and significant improvements in core functionality, making data review and acquisition easier and more intuitive for users. “Key enhancements were made to the instrument set-up and deconvolution areas of this mass spectrometry data system,” said Lorne Fell, Product Manager for Separation Science. “We conducted a global Beta program and based our improvements and additions on the feedback we received. Some of the tools developed included advanced filters and compound class-specific mass defect plots so users can easily review their most relevant data.” In addition, Fell says, the use of automated formula matching for deconvoluted fragment ions and ChemSpider to help identify “unknown/unknowns” give users the best means to evaluate their results fully and efficiently. ChromaTOF-HRT 1.81 also includes a number of acquisition centric upgrades with a focus on ease-of-use. These features include a quick run button, the ability to conduct a Tune Report, and significant improvements to both the auto-tune and gain optimization algorithms. The software now also supports the use of hydrogen as a carrier gas. The same great features of the previous software version continue to be available with ChromaTOF-HRT 1.81, including the newly termed High Resolution Deconvolution™ (HRD™) for creation of pure component mass spectra, seamless data handling, and full compatibility with the NIST library to identify known/unknowns. Overall, the release of ChromaTOF-HRT version 1.81 allows users to be easily successful in gathering and analyzing data from the GC-HRT, and allowing them more time to answer the truly difficult chemical questions in their labs. About LECO Corporation For over 75 years, industries around the world have trusted LECO Corporation to deliver technologically advanced products and solutions. Today, that commitment continues with high-speed/high resolution Time-of-Flight Mass Spectrometry (TOFMS) as well as comprehensive two-dimensional gas chromatography (GCxGC), all featuring easy-to-use ChromaTOF® operating software. Product lines also include high-quality analytical instrumentation, metallography and optical equipment, and consumables. LECO currently has over 30 subsidiaries worldwide, in countries such as the United Kingdom, Australia, Sweden, Canada, and Mexico, with additional distributors authorized to sell or service LECO products to the rest of the world. For more information, visit www.leco.com, or follow us on Twitter (@LECOCorp), and LinkedIn. Source: LECO Corporation Press Release |
Metabolomics Events
|
| 29 Jan 2015 |
Live webinar: Dr. Oscar
Yanes - "Metabolomics: only suitable for
multidisciplinary teams" Session
1: Expert Speaker, 20 min
presentation followed by 10 min Q/A Metabolomics
is defined as the comprehensive and
quantitative analysis of metabolites in living
organisms. Among the omic sciences,
metabolomics is possibly the most
multidisciplinary of all, involving knowledge
about electronic engineering and signal
processing, analytical and organic chemistry,
biostatistics and statistical physics, and
biochemistry and cell metabolism. Here an
untargeted metabolomics workflow will be
detailed that provides examples of this
multidisciplinarity. Oscar Yanes has long experience in developing
new technologies, methods and applications in
mass spectrometry-based metabolomics. Recently
he is also implementing NMR metabolomic
approaches to study fundamental biological
processes in combination with other omic
platforms. He has significant publications to
his name in high-impact factor journals
including Nature, Nature Chemical Biology and
Journal of Clinical Investigation, with more
than 1,000 citations to date. Registration: https://attendee.gotowebinar.com/register/2409752719256762369 |
| 9-12 Feb 2015 |
3rd Annual Practical
Applications of NMR in Industry Conference
(PANIC) The PANIC meeting is intended to address topics that occur daily in industrial, government, and academic research laboratories whose primary task entails the application of NMR to a diverse set of analytical problems. Topics include quantitation, molecular structure characterization, trace component and mixture analysis, and product support for a variety of materials that include small molecules, polymers, heterogeneous mixtures, natural products, biosimilars, polysaccharides, and proteins. The conference provides in-depth discussions of the “nuts and bolts” of basic NMR experiments that accent the underlying best-practices developed to address these everyday problems. It also explores the regulatory aspects of the applications of these experiments. Greater insights will be provided into the less frequently explored techniques used in NMR such as quantitation, chemometrics, automation, relaxometry, and at/on line instrumentation. Attendees will have the opportunity to learn about and discuss applications of NMR to polymers, petroleum, food, agriculture, and nutritional supplements that are rarely discussed at other NMR meetings. Ample opportunity will be given to Network with people who are experts in inventing NMR experiments to solve real world problems in a timely and efficient manner that is demanded by working in the venue of product delivery. For more details, visit http://www.panicnmr.com. |
| 16-20 Feb 2015 |
EMBO Practical Course on
Metabolomics Bioinformatics for Life
Scientists Organizers
Participation: Open application
with selection |
| 28 Mar to 1 Apr 2015 |
Plant Metabolism Session
at the ASBMB Annual Meeting |
| 15-16 Jun 2015 |
Informatics and
Statistics for Metabolomics (2015) A poster announcing this workshop can be found here.The workshop will cover many topics ranging from understanding metabolomics technologies, data collection and analysis, using pathway databases, performing pathway analysis, conducting univariate and multivariate statistics, working with metabolomic databases and exploring chemical databases. Participants will be given various data sets and short assignments to assist with the learning process. Target Audience This course is intended for graduate students, post-doctoral fellows, clinical fellows and investigators who are interested in learning about both bioinformatic and cheminformatic tools to analyze and interpret metabolomics data. Prerequisite: Familiarity with R is required. Familiarity can be gained through online activities. You should be familiar with these R concepts (chapters 1-5) or review the past Statistics tutorials provided by CBW. Apply Now Award Opportunities For further details, visit http://bioinformatics.ca/workshops/2015/informatics-and-statistics-metabolomics-2015. |
| 29 Jun to 2 Jul 2015 |
Metabolomics 2015: 11th
Annual Conference of the International
Metabolomics Society You are invited to join us for Metabolomics
2015, the official annual meeting
of the Metabolomics Society. For further details, visit http://metabolomics2015.org. |
Metabolomics Jobs |
This is a resource for
advertising positions in metabolomics. If you have a
job you would like posted in this newsletter, please
email Ian Forsythe (metabolomics.innovation@gmail.com).
Job postings will be carried for a maximum of
4 issues (8 weeks) unless the position is filled prior
to that date.
Jobs
Offered
| Job Title | Employer | Location | Posted | Closes | Source |
|---|---|---|---|---|---|
| Postdoctoral
Fellow in Environmental Metabolomics |
University
of Toronto |
Toronto, Canada | 8-Jan-2015 |
28-Feb-2015 |
naturejobs.com |
| Postdoctoral
scientist in metabolomics |
MRC
National Institute for Medical Research
/ Francis Crick Institute |
London, UK | 5-Jan-2015 | 20-Jan-2015 |
Francis
Crick Institute |
| Mass
Spectrometry Metabolomics Postdoc at the MRC
Regional Phenome Centre |
University
of Birmingham |
Birmingham, UK | 23-Dec-2014 | 25-Jan-2015 |
University
of Birmingham |
| Vice
Chancellor's Postdoctoral Fellowship |
RMIT
University |
Melbourne, Australia | 18-Dec-2014 |
8-Feb-2015 |
RMIT
University |
| Vice
Chancellor's Research & Senior Research
Fellowships |
RMIT
University |
Melbourne, Australia | 18-Dec-2014 |
8-Feb-2015 |
RMIT University |
| Principal
Statistician |
UC Davis |
Davis, California, USA | 18-Dec-2014 | Open until filled |
UC
Davis |
| Research
Engineer in quantitative mass spectrometry |
MetaboHUB -
INRA |
Bordeaux, France | 17-Dec-2014 | 15-Jan-2015 |
MetaboHUB |
| Assistant/Associate
Professor, Environmental Health Sciences -
Metabolomics |
Yale
University |
New Haven, USA | 9-Dec-2014 | 15-Jan-2015 |
naturejobs.com |
| Postdoctoral
Researcher in Metabolomics of Cancer |
Karolinska
Institutet |
Stockholm, Sweden | 27-Nov-2014 | 31-Jan-2015 |
Karolinska
Institutet |
|
Ian
J. Forsythe, M.Sc.
MetaboNewsEditor Department of
Computing Science
University of Alberta 221 Athabasca Hall Edmonton, AB, T6G 2E8, Canada Email: metabolomics.innovation@gmail.com Website: http://www.metabonews.ca LinkedIn: http://ca.linkedin.com/in/iforsythe Twitter: http://twitter.com/MetaboNews Google+: https://plus.google.com/118323357793551595134 Facebook: http://www.facebook.com/metabonews |
This newsletter
is published in partnership between The
Metabolomics Innovation Centre (TMIC, http://www.metabolomicscentre.ca/)
and the Metabolomics Society (http://www.metabolomicssociety.org)
for the benefit of the worldwide
metabolomics community.
 A single source destination for fee-for-service metabolic profiling including comprehensive metabolite identification, quantification, and analysis 
|