The False Positive Paradox in Metabolomics
Feature article contributed by Caroline Johnson, Winnie
Uritboonthai, and Gary
Siuzdak,The Scripps Research Institute, La Jolla,
California
The False Positive Paradox is defined as a statistical outcome
where a false positive is more probable than a true positive.
Metabolite identification represents an example of the paradox,
especially when accurate mass measurements alone are used for
identification. An extreme example would be searching METLIN (
metlin.scripps.edu) with a
neutral mass of 180.0634 (the mass of glucose), this particular
entry produces 24 hits within 2 parts per million (ppm)
accuracy.
Analytical Chemistry recently highlighted an article from
our lab that discussed the importance of tandem mass spectrometry
analysis (
Figure
1,
Patti,
G.J. et al. Anal. Chem. 2013, 85(2):798-804) in
metabolite characterization, as well as the introduction of XCMS
Cloud Plots (
http://xcmsonline.scripps.edu/).
Specifically, this article compared the use of precursor accurate
mass measurements by themselves for identifying metabolites versus
using precursor and tandem mass spectrometry data. The purpose of
the plot was to illustrate the level of false positives that come
from using just accurate mass measurements of precursor ions for
identification purposes.
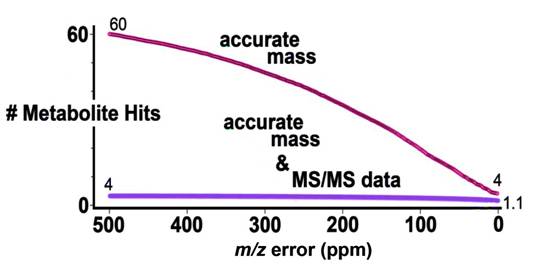 Figure 1.
Figure 1. The number of metabolite hits generated from the
METLIN data base as a function of m/z error (top) and m/z error
and tandem mass spectrometry data. Tandem mass spectrometry data
offers significantly higher confidence and lower rate of false
positives.
It is intuitively obvious that metabolite
assignments based on accurate mass measurements alone create a
high level of false positive identifications. Two reasons for
this redundancy are the existence of isobaric metabolites
(compounds with identical mass) and secondly, metabolites that
have similar masses that cannot be resolved with the mass
accuracy of many mass analyzers. To illustrate this, the number
of metabolite hits from the METLIN database was plotted as a
function of accurate mass measurements and mass error, and a
second superimposed plot calculated the number of hits based on
a combination of accurate mass measurements, tandem mass
spectrometry (MS/MS) data, and mass error (Figure
1). Using this plot, even at the extreme of accuracy
(0.0 ppm error) an average of 4 metabolites were identified
based on accurate mass measurements alone, hence the reference
to the “paradox”. However when accuracy was combined with
tandem mass spectrometry data, the average number of
identifications dropped to 1.1.
A specific example of the false positive paradox
is shown in Figure
2 with data on three isobaric
metabolites that have identical elemental composition and
mass, and therefore are indistinguishable based on accurate
mass measurements. It
is only possible that, at best, one of these three is correct.
Fortunately, as shown,
the compounds have distinct tandem fragmentation patterns, one
of which matched to a key metabolite implicated in neuropathic
pain (Patti,
G.J. et al., Nat. Chem. Biol. 2012, 8(3):232-4).
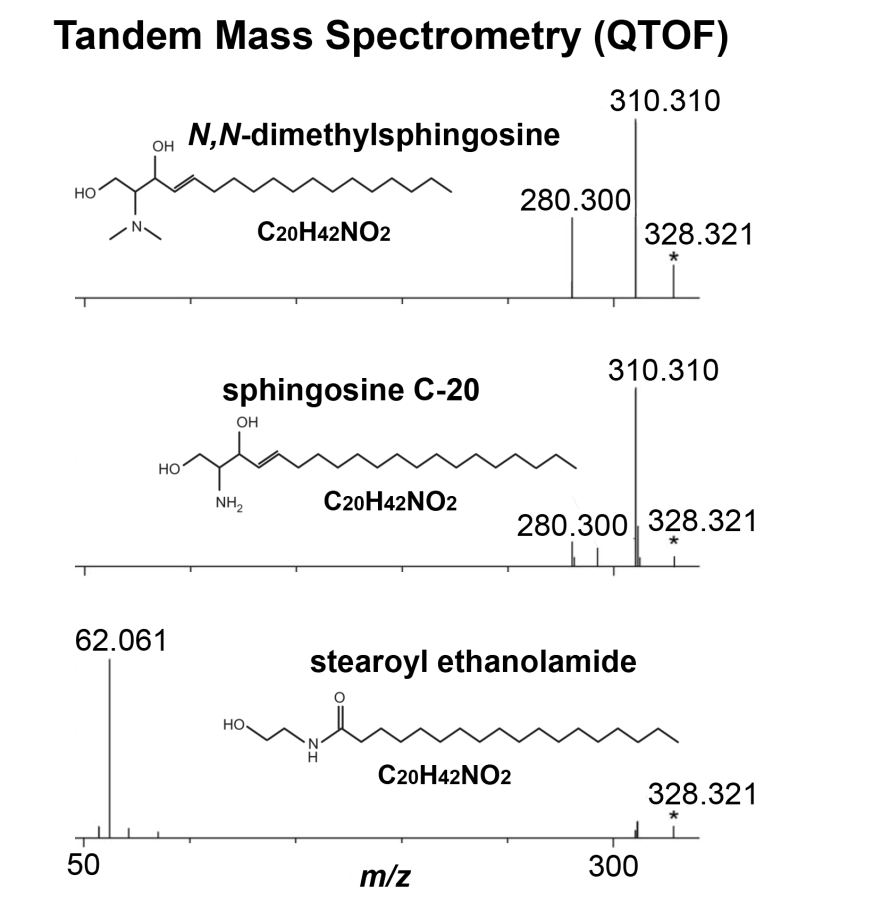
Figure 2. Tandem mass spectrometry data
used to distinguish dimethylsphingosine (DMS), sphingosine
C-20, and stearoyl ethanolamide, all having the same elemental
composition. Each of these metabolites has the exact same
molecular weight and are therefore indistinguishable based on
accurate mass measurements. The importance of peak intensity
should also be noted, for example, using in silico
measurements (without accurate experimental intensities), DMS
and C-20 would be indistinguishable.
In order to at least minimize the false positive
paradox in metabolomics and improve the assignment statistics
behind metabolite identification, we've created the METLIN
database with over 75,000 molecules and 56,000+ tandem mass
spectra. So while with
mass spectrometry technology it is possible to observe many
peaks (features) corresponding to a wide range of metabolites,
given the high redundancy in the mass of metabolites, accurate
mass alone is incapable of identifying them. However, tandem
mass spectrometry will not completely eliminate false
positives especially in cases of stereoisomers, or lipids
where the location of double bonds is not well defined,
nonetheless it is a step closer to identifying metabolites
with a higher level of confidence.
On a separate yet related note, I would like to
acknowledge my gratitude to the companies, institutions, and
laboratories that have donated and continue to provide us with
compounds that allow us to generate the tandem mass
spectrometry data in METLIN. They
include
SIGMA-Aldrich, Cayman Chemical, ChromaDex, Joint BioEnergy
Institute (JBEI), Millenium Laboratories, Genomics Institute
of the Novartis Research Foundation (GNF), UCSD (William
Gerwick), TSRI (Dale Boger), and the NIH/NCI. We are also open
to any new metabolic entities to add to METLIN, whether they
are of plant, animal, or micro-organism origin. If you have metabolites that
you wish to contribute, please contact us (see the contact
information below).
METLIN Contact Information
Please
note: If you know of any
metabolomics research programs, software, databases,
statistical methods, meetings, workshops, or training
sessions that we should feature in future issues of this
newsletter, please email Ian Forsythe at metabolomics.innovation@gmail.com.









