Metabolite NMR Signals Show Quantification Differences When
Comparing Different NMR Probes and Parameters
Paige Lacy,1 Ryan T. McKay,2 Michael
Finkel,3 Alla Karnovsky,4 Scott Woehler,5
Michael J. Lewis,6 David Chang,6 and
Kathleen A. Stringer3,4
1Pulmonary Research Group, Department of Medicine
and
2Department of Chemistry, University of Alberta,
Edmonton, Alberta, Canada;
3Department of Clinical
Sciences, College of Pharmacy,
4Department of
Computational Medicine and Bioinformatics, School of Medicine,
5Department
of Medicinal Chemistry and the Biochemical Nuclear Magnetic
Resonance Core, College of Pharmacy, University of Michigan, Ann
Arbor, MI, USA; and
6Chenomx, Edmonton, Alberta,
Canada
Metabolomic measurement of human samples is fast becoming a
priority for development of novel diagnostic biomarkers of
disease. Many metabolomic centres are becoming established around
the world to quantify metabolites in human samples using NMR- and
mass spectrometry-based approaches, and substantial funding for
these is being provided by the NIH and other major granting
agencies.
Human urine samples are a prime choice for metabolomic analysis
because of their ease of collection and analysis. NMR-based
metabolomics is one of the best approaches for the analysis of
complex biological samples such as urine, as very little sample
preparation is required, and it provides remarkably robust
biomarker candidates for many diseases. The reproducibility and
validity of NMR measurements has not been compared across
different sites for a given sample population. We discovered that
serious issues arose that complicated interpretation of
metabolomic data, even when identical samples were analyzed.
This startling observation arose from a collaboration with Drs.
Kathleen Stringer and Alla Karnovsky at the University of Michigan
to determine the reproducibility of NMR measurements of split
human urine samples at two different sites. Technical replicates
of urine samples were assayed by 1D-
1H-NMR at both the
University of Alberta and the University of Michigan. Urine
samples were obtained from healthy volunteers at the University of
Michigan under a standard operating procedure for collection and
processing.
Our initial goal was to verify that comparable data could be
acquired at more than one NMR facility. Not surprisingly, we
obtained comparable quantifications for many molecules within
samples, since these derived from exactly the same sample. But
what we quickly observed was that, by using even slightly
different NMR parameters on the two instruments, we obtained
significantly different concentrations for some metabolites within
the same sample. Subsequent analysis using standard statistical
techniques revealed that quantitative data across sites can be
achieved, but (disturbingly) unrecognized NMR parameter
differences had some dramatic and widely perturbing effects on the
results. One particular parameter showing marked differences was
the use of 3 mm vs. 5 mm probes, which showed varying degrees of
solvent suppression (see Figures
1
and
2).
While 5 mm probes at both sites showed similar spectral results,
the 3 mm probes consistently showed marked solvent suppression,
with peaks sometimes disappearing near the water peak. This has an
obvious impact on the identification and quantification of urinary
metabolites.
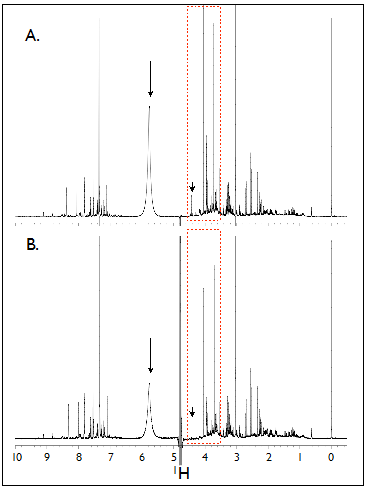 Figure 1. Comparison of 1H-NMR spectra of
human urine samples using identical parameters settings without
calibration.
Figure 1. Comparison of 1H-NMR spectra of
human urine samples using identical parameters settings without
calibration. NMR spectra collected from the same human
urine sample using a 5 mm probe (A) and a 3 mm probe (B). Note the
appearance of a peak in (A) which disappears in (B) (arrow in red
dashed box). (Figure from Lacy P
et al., Signal
intensities derived from different NMR probes and parameters
contribute to variations in quantification of metabolites,
PLoS
One. 2014 Jan 21;9(1):e85732. doi:
10.1371/journal.pone.0085732.
eCollection 2014.)
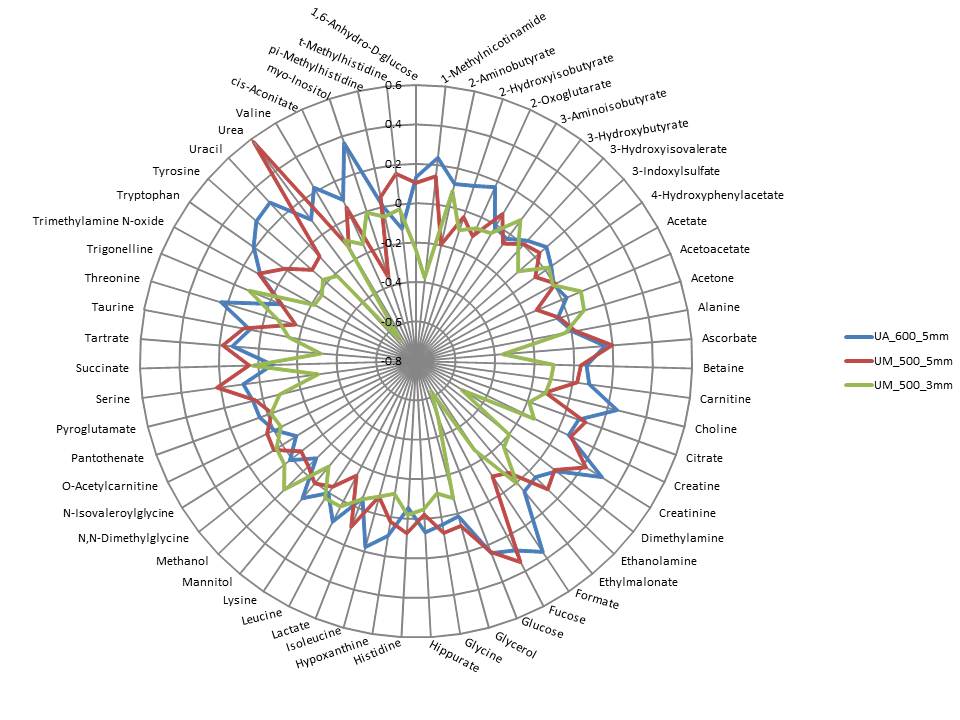 Figure 2. Comparison of 1H-NMR-measured metabolites
in human urine samples using 3 and 5 mm probes.
Figure 2. Comparison of 1H-NMR-measured metabolites
in human urine samples using 3 and 5 mm probes. This radial
plot shows the variation in levels of metabolites using 3 and 5 mm
probes at the Universities of Alberta and Michigan. The greatest
variations occur when comparing the 3 mm probe data with that of 5
mm data. (Figure from Lacy P
et al., Signal intensities
derived from different NMR probes and parameters contribute to
variations in quantification of metabolites,
PLoS One.
2014 Jan 21;9(1):e85732. doi:
10.1371/journal.pone.0085732.
eCollection 2014.)
The implications of this observation are significant and
problematic, as cross-center validation metabolomics studies are
essential for the reliable application of metabolomics to
biomarker discovery and clinical applications. This poses similar
issues for single site studies using a single spectrometer for all
data acquisition. It was realized that not only do NMR parameters
have to be maintained, but also that the subsequent performance of
the instrumentation must be consistent over time, or has to be
regularly calibrated, which is not consistently done in our
experience.
We presented in our paper (
Lacy
et al. (2014) PLoS One, DOI:
10.1371/journal.pone.0085732, published January 21, 2014) a
confirmed validation of NMR analysis at two sites, and report the
range and magnitude that common NMR parameters involved in solvent
suppression had on quantitated metabolomics data. A specific
example was that saturation power levels greatly influenced peak
height intensities in a frequency-dependent manner for a number of
metabolites, which markedly impacted the quantification of
metabolites. This is to say that metabolite concentrations were
increasingly inaccurate closer to the solvent signal.
Collectively, these findings highlighted the importance of and
need for exceedingly consistent use of precisely calibrated NMR
parameter settings within and across centers in order to generate
reliable, reproducible quantified NMR metabolomics data. For any
studies associated with NMR analysis of urine samples, a
calibration profile should be established using standardized
samples to determine the envelope of saturation effect using the
probe and spectrometer for the individual studies.
Please
note: If you know of any
metabolomics research programs, software, databases,
statistical methods, meetings, workshops, or training
sessions that we should feature in future issues of this
newsletter, please email Ian Forsythe at metabolomics.innovation@gmail.com.









