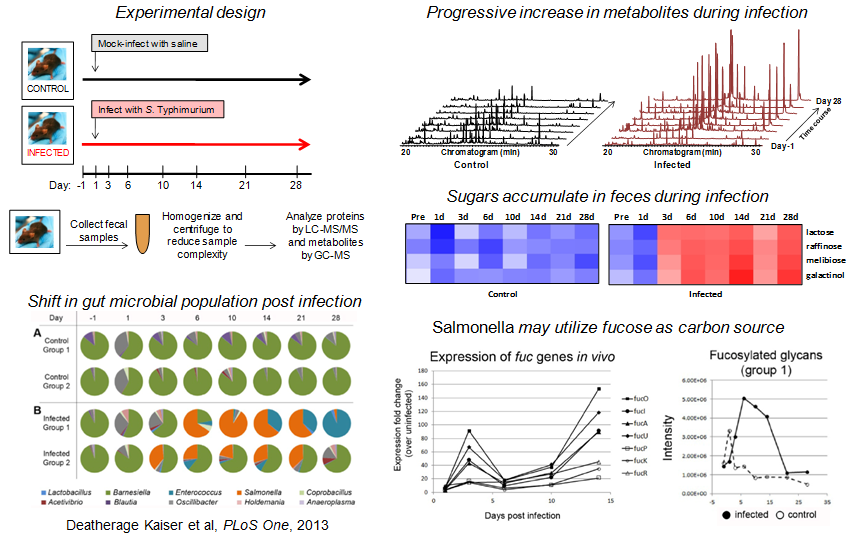
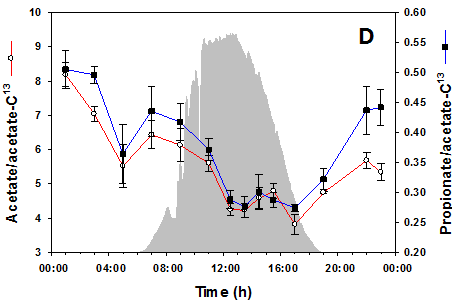
MxP® Quality
Control Plasma—a
metabolomics-based biomarker for holistic human plasma quality
control
"Unacceptable sample quality due to failures in the
pre-analytical phase and/or improper sample storage in
retrospective research studies is a widely known challenge in
biomedical R & D."
Prof. Dr. Kurt Zatloukal, Coordinator of a
pan-European Research Infrastructure on Biobanking and
Biomolecular Resources - BBMRI
Feature article contributed by Dr. Beate Kamlage,
Senior Scientist, Metanomics GmbH, Tegeler Weg 33, 10589
Berlin, Germany
Any clinical research and development study depends on
high-quality specimen collection to hunt for new drug targets and
biomarkers. Pre-analytical processing steps of blood including
choice of sample tube, prolonged storage of blood or plasma and
incubation at improper temperatures can have negative effects on
biomolecules. Therefore, both quality assurance (e.g., definition
of stringent SOPs) and sample quality control (QC), achieved
through quality control biomarkers are important concepts to
ensure the validity of clinical results.
Metabolite levels in blood cover a wide range of physiological and
chemical processes, and are very reactive to disturbances.
Analysis of the responsiveness of the human plasma metabolome to
pre-analytical variations is allowing comprehensive and sensitive
read-out of sample quality. In other words, it tells researchers
about the actual condition of their plasma samples. This
information is especially essential for omics-based biomarker
research as well as for system biology approaches and biobanking.
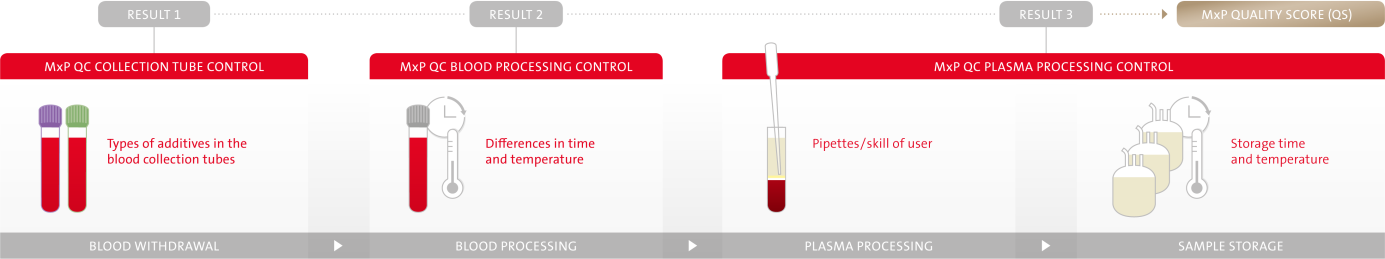
Figure 1. Common confounders in the pre-analytical phase of plasma
are covered by
MxP® Quality Control Plasma by delivering 3 test
results.
MxP® Quality
Control Plasma
is a novel, validated, metabolome-based assay by Metanomics Health
GmbH, which, for the first time, provides a holistic quality check
of EDTA human plasma samples by delivering three test results.
These results provide comprehensive information about presence of
pre-analytical confounders and where they have occurred in the
process (Figure 1). This enables researchers to upgrade their
quality management (QM) system from quality assurance to quality
control, safe-guarding their investment in the right human plasma
samples.
Example A. MxP® Quality Control Plasma detecting batch effects
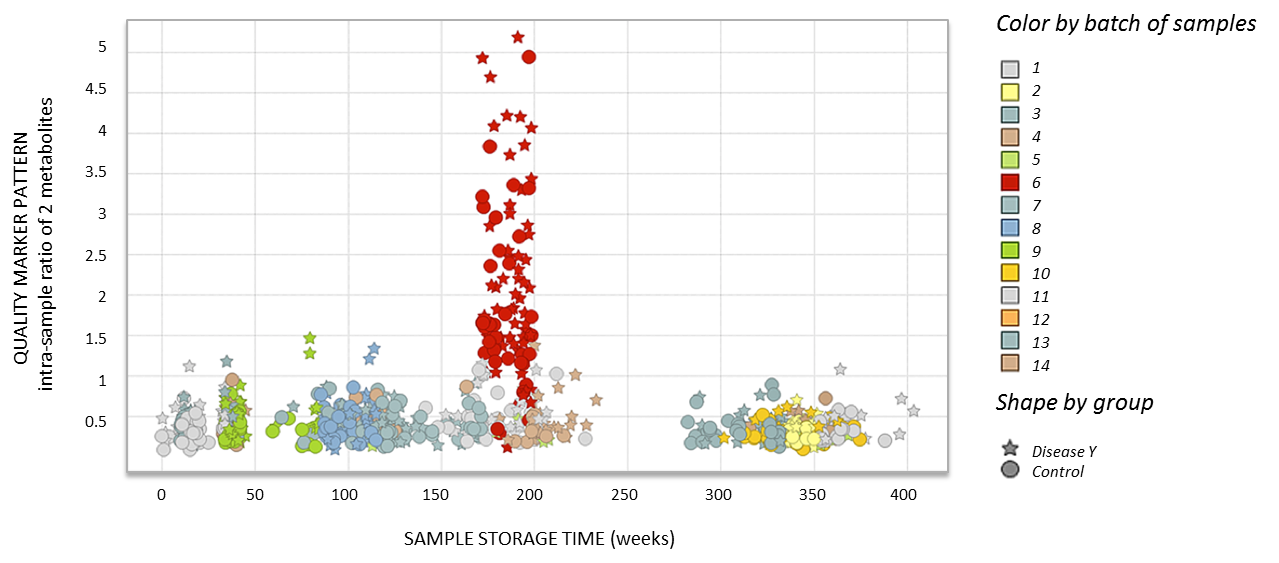
Figure 2. Metabolic quality marker pattern indicating a
pre-analytical issue related to plasma processing in a batch of
samples (red) from a clinical biomarker study.
The importance of quality control in addition to quality assurance
for biobanking and multi-center clinical trials is concluded from
the example shown in Figure 2. All samples in this study
(represented by dots and stars) were taken from the same biobank,
which received them from various clinical centers. We noticed a
particular group of samples (marked in red) with an increase in
the metabolite pattern, detected by Metanomics Health, which
represents the low quality of the specimens. When specific colors
were assigned to every batch of samples, it appeared that all
outlying results correspond to the same batch. Every batch
represents samples that have been taken out of the biobank at the
same time point, thawed, split into aliquots, and shipped. The
overall quality of the samples, represented by low intra-sample
ratio, indicates that a very specific pre-analytical issue must
have occurred in this particular batch due to non-conformant
processing conditions. This can be identified only by a quality
control service as SOPs (standard operating procedures) were
claimed to be followed for all the samples in the study.
Example B. MxP® Quality Control Plasma improving the outcome of
clinical R&D and biomarker research
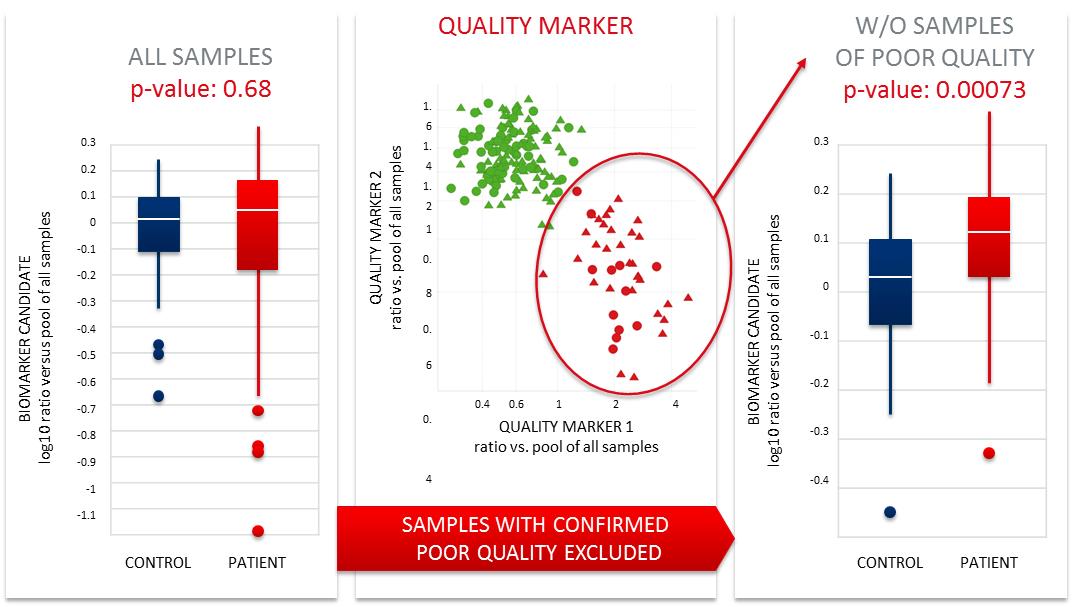
Figure 3. Example of a beneficial effect of sample quality control
in a biomarker identification study.
Figure 3 illustrates the beneficial effect of sample quality
control in clinical biomarker research. As shown on the left side,
initially no significant difference (p-value>0.05) between the
disease and control group was found. By additional quality
assessment of the samples, a sub-group of samples of poor quality
(marked in red) could be identified and removed. As shown on the
right side, the statistical analysis, repeated on the set of
satisfactorily qualified samples, elucidated a significant
difference (p-value<0.05) for the same biomarker candidate and
additionally a lower variability of the two groups (see the
vertical axis). This example demonstrates the importance of
quality control assessment prior to conducting research.
Investing in the right samples
The novel
MxP® Quality Control Plasma
assay measures different validated metabolomic biomarkers in
single read-outs. By applying proprietary algorithms, three
different test results are calculated, which provide not only a
precise assessment of sample quality but also a “fit for purpose”
suggestion for sample utilization.
This is enabling pharmaceutical R&D, clinical research
organizations, and biobanks to better understand the actual
condition of human plasma samples, efficiently monitor SOP
compliance in multicenter trials, deliver superior quality
samples, and support evidence-based decisions for sample
selection.
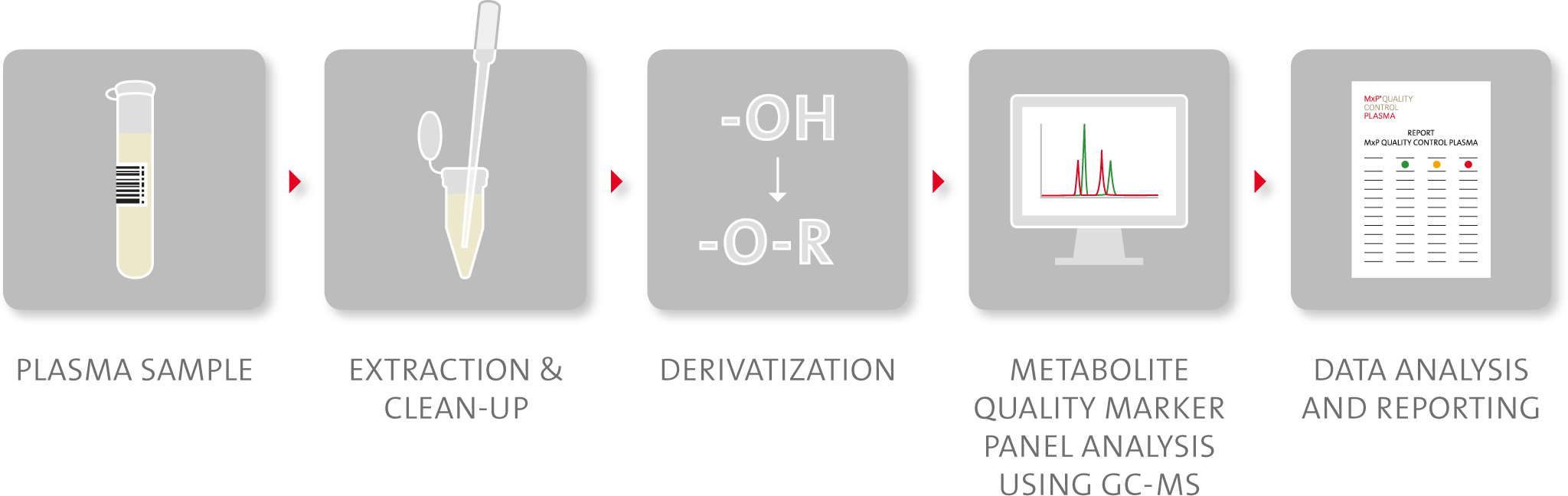
Figure 4. Schematic representation of
MxP®
Quality Control Plasma workflow.
Details of the plasma metabolome response to pre-analytical
confounders have been recently published in a peer-reviewed
article (“Quality Markers Addressing Preanalytical Variations of
Blood and Plasma Processing Identified by Broad and Targeted
Metabolite Profiling”, Clinical Chemistry, 2013,
http://www.clinchem.org/content/60/2/399.full).
Metanomics Health offers its quality control assay for human
plasma samples in a fee-for-service and an out-licensing business
model.
Metanomics Health, a BASF Group company, is the world-leading
company offering targeted and non-targeted metabolomics to
healthcare, nutrition, and bioprocessing partners in industry and
academia. In parallel to the service business, Metanomics Health
is funding and conducting a comprehensive clinical biomarker
program, addressing a wide range of questions with high unmet
medical need.
Find out more about Metanomics Health GmbH at www.metanomics-health.de
Please
note: If you know of any
metabolomics research programs, software, databases,
statistical methods, meetings, workshops, or training
sessions that we should feature in future issues of this
newsletter, please email Ian Forsythe at metabolomics.innovation@gmail.com.