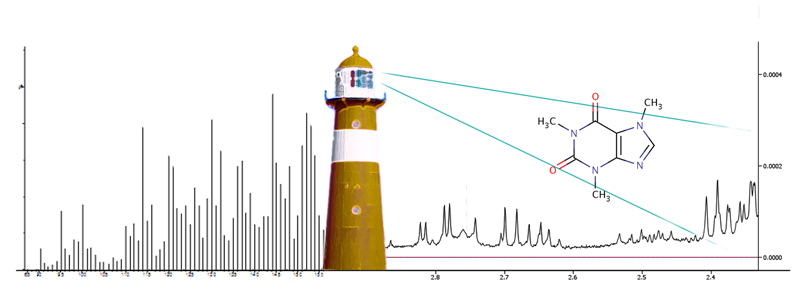
MetaboLights - The new EBI Metabolomics database
Article
contributors: Kenneth Haug1, Dr. Reza M. Salek1,3,
Pablo Conesa1,
Tejasvi Mahendraker1, Dr. Mark Williams1,
Dr. Julian L. Griffin2,3, Dr. Christoph
Steinbeck1
Affiliations:
1. Cheminformatics and Metabolism Group, European
Bioinformatics Institute, Wellcome Trust Genome Campus, Hinxton,
Cambridgeshire, CB10 1SD, UK.
2. MRC HNR, Elsie Widdowson Laboratory, Fulbourn Road,
Cambridge, CB1 9NL, UK.
3. University of Cambridge, Department of Biochemistry and
Cambridge Systems Biology Centre, Cambridge CB2 1GA, UK.
Over the past ten years a significant number of databases for
Metabolomics have been created, differing in the types of data,
species and applications they cover. Recently, efforts are
underway to establish large-scale repositories for metabolomics
data as they exist for the other omics areas, both in Europe as
well as in the United States.
In June 2012, EMBL-European Bioinformatics Institute (EBI),
Europe's largest open-access data provider in molecular biology,
launched MetaboLights (
http://www.ebi.ac.uk/metabolights),
the first general-purpose, open-access repository for metabolomics
studies, their raw experimental data and associated metadata. The
EBI has a proven record of maintaining such databases and its
associated data over several decades, thereby ensuring long-term
stability and exploitability.
The MetaboLights repository [
1],
powered by the open source ISA framework [
2],
is cross-species and cross-technique. MetaboLights features an
open repository layer (released in June 2012 and receiving data)
and ultimately a curated knowledge base/reference layer with
abstracted information on metabolites, their chemistry, biology,
and analytical data. For the MetaboLights repository, scientists
submit information about a metabolomics study, comprising the
primary research data and associated meta-data. Studies
automatically receive a stable unique accession number that can be
used as a publication reference (e.g., MTBLS1,
Figure
1).
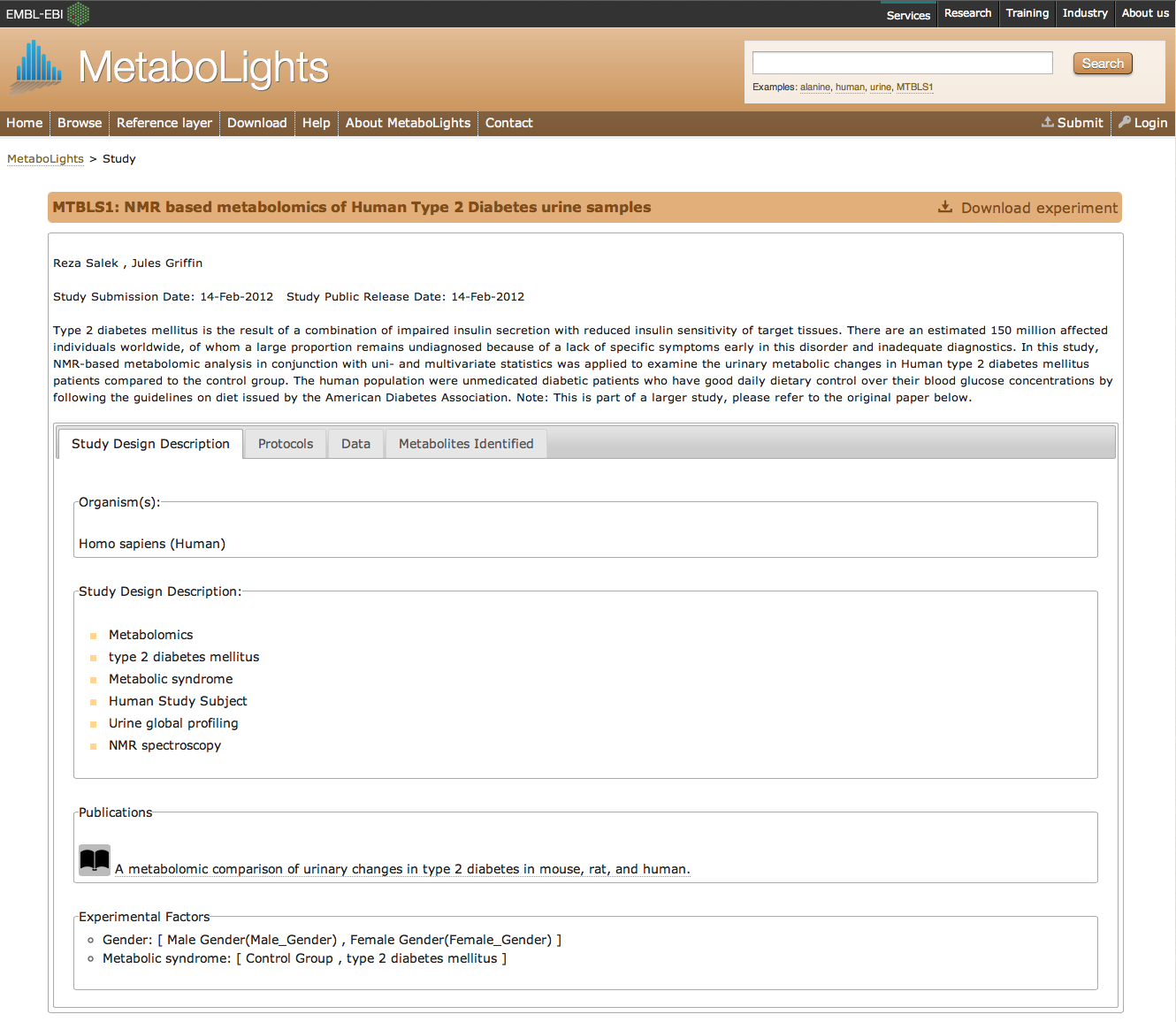
Figure 1. A screenshot of study MTBLS1 - NMR based
metabolomics study on Type 2 Diabetes.
MetaboLights accept studies in ISA-Tab format [
3].
ISA-Tab is rapidly becoming the
de facto standard for
bioscience data exchange.
In October 2012 the Cheminformatics and Metabolism team at the EBI
started the global COSMOS [
4]
project (Coordination of Standards in Metabolomics) (
www.cosmos-fp7.eu). COSMOS
will bring together European data providers to set and promote
community standards that will make it easier to disseminate
metabolomics data through life science e-infrastructures.
The MetaboLights website provides a pre-configured ISAcreator for
creating and editing the ISA-Tab files (
http://www.ebi.ac.uk/metabolights/download)
(
Figure
2). This archive can be downloaded and you have all you need
to start recording your experiments. MetaboLights will encode
agreed standards from the COSMOS project to facilitate high
quality data exchange.
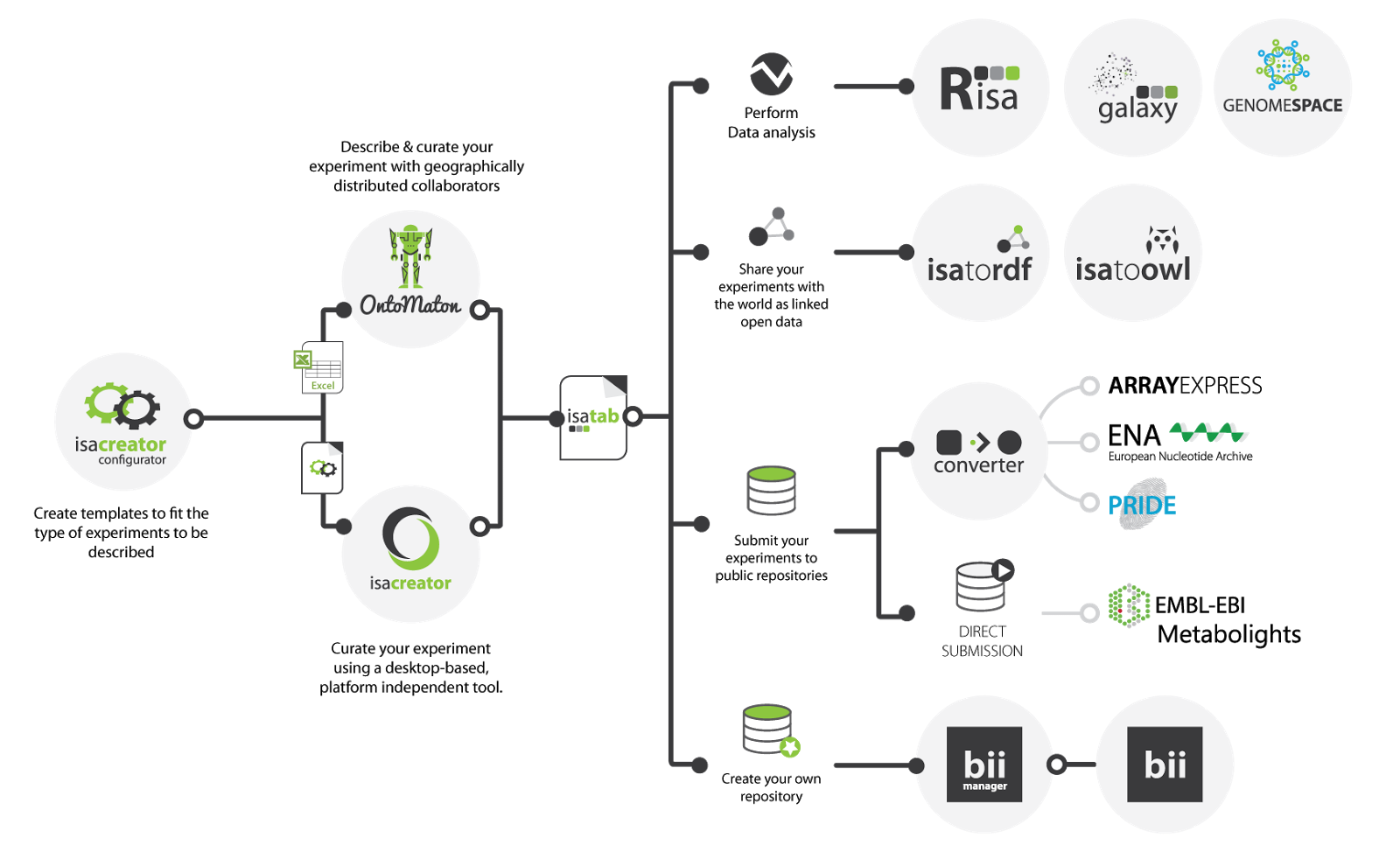
Figure 2. Workflow for the ISA software suite, with direct
submission to MetaboLights.
To simplify sharing and reporting of any metabolites found in a
study, we have developed a metabolite identification plugin to
ISAcreator (
Figure
3). This plugin will search for additional compound
information from PubChem.
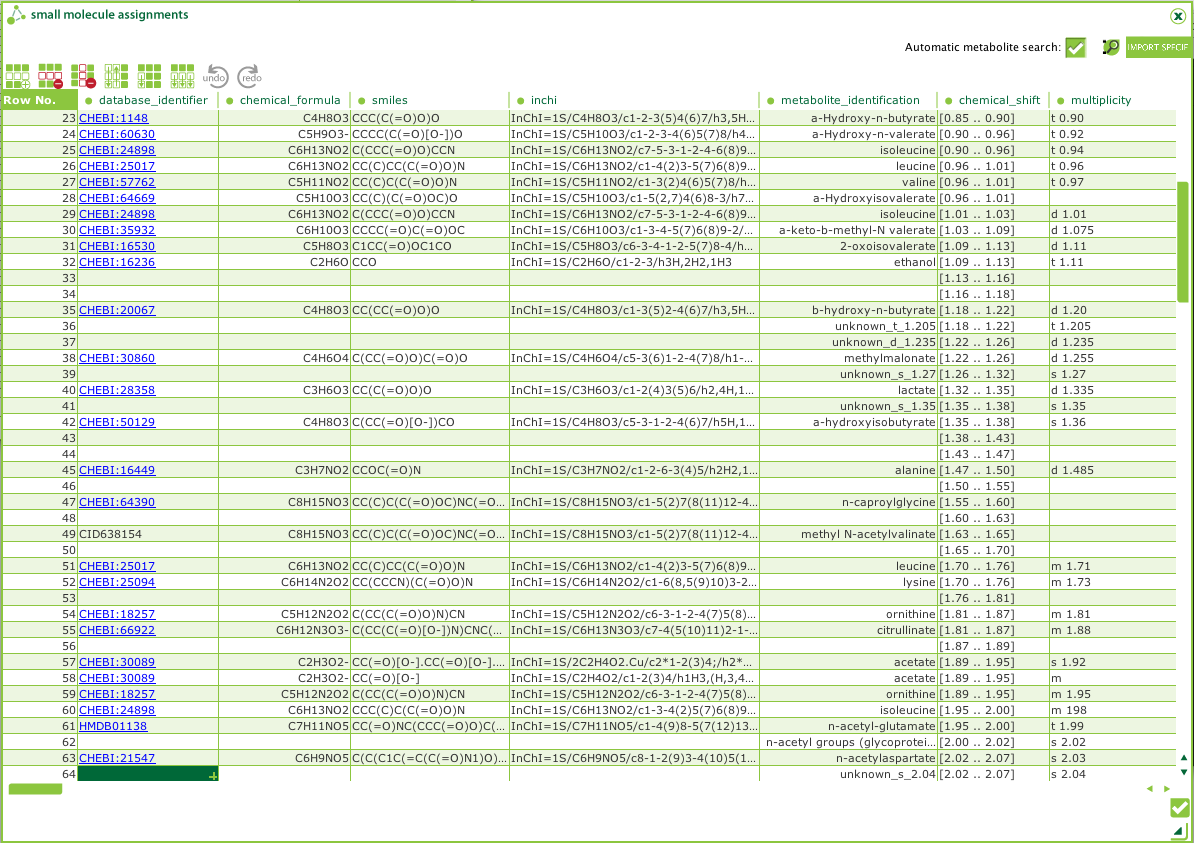 Figure 3.
Figure 3. MetaboLights - ISAcreator Metabolite
Identification Plugin.
At present, the repository includes 22 submitted studies,
encompassing 152 protocols for 5253 samples, and span over ten
different species including human,
Caenorhabditis elegans,
Mus musculus, and
Arabidopsis thaliana. As many as
937 of the metabolites identified in these studies have been
mapped to ChEBI (
http://www.ebi.ac.uk/chebi)
and 135 to HMDB (
http://www.hmdb.ca).
These studies cover a variety of techniques, including NMR
spectroscopy and mass spectrometry.
The MetaboLights team are now actively implementing the
MetaboLights Reference Layer, which will be launched in Summer
2013. This will be a metabolomics knowledge base with the
metabolite as the central element. This will enable
experimentalists to get a comprehensive Metabolomic view on known
metabolites. There will be information about the reference
biology, metabolites and their occurrence and concentration in
species, organs, tissues, and cell compartments in various
conditions e.g., healthy vs diseased.
A short introduction to MetaboLights is now available on
EBI
Train online.
References
- Kenneth Haug, Reza M. Salek, Pablo Conesa, Janna Hastings,
Paula de Matos, Mark Rijnbeek, Tejasvi Mahendraker, Mark
Williams, Steffen Neumann, Philippe Rocca-Serra, Eamonn
Maguire, Alejandra Gonzalez-Beltran, Susanna-Assunta Sansone,
Julian L. Griffin and Christoph Steinbeck. MetaboLights--
an open-access general-purpose repository for metabolomics
studies and associated meta-data. Nucl. Acids Res.
(2012). [doi: 10.1093/nar/gks1004]
- Sansone, S.A., Rocca-Serra, P., Field, D., Maguire, E.,
Taylor, P., Hofmann, O., Fang, H., Neumann, S., Tong, W.,
Amaral-Zettler, L., et al. (2012). Toward
interoperable bioscience data. Nature Genetics
44, 121-126. [doi:10.1038/ng.1054]
- Rocca-Serra, P., Brandizi, M., Maguire, E., Sklyar, N.,
Taylor, C., Begley, K., Field, D., Harris, S., Hide, W.,
Hofmann, O. et al. (2010). ISA
software suite: supporting standards-compliant experimental
annotation and enabling curation at the community level.
Bioinformatics, 26, 2354-2356.
[doi:10.1093/bioinformatics/btq415]
- Christoph Steinbeck, Pablo Conesa, Kenneth Haug, Tejasvi
Mahendraker, Mark Williams, Eamonn Maguire, Philippe
Rocca-Serra, Susanna-Assunta Sansone, Reza M. Salek, Julian L.
Griffin. MetaboLights:
towards a new COSMOS of metabolomics data management. Metabolomics,
October 2012, Volume 8, Issue 5, pp 757-760.
[doi:10.1007/s11306-012-0462-0]
Please
note: If you know of any
metabolomics research programs, software, databases,
statistical methods, meetings, workshops, or training
sessions that we should feature in future issues of this
newsletter, please email Ian Forsythe at metabolomics.innovation@gmail.com.