Metabolomics Australia and its Bioinformatics Capabilities
Feature article contributed by Saravanan Dayalan1,
Seán O'Callaghan1, David P DeSouza1,
Brad Power3, Tamas Szabo3, Adam Hunter3,
Matthew Bellgard3, Dedreia L Tull1, Ute
Roessner2, Vladimir A Likic1, Malcolm J
McConville1, and Antony Bacic1,2
1 Metabolomics Australia, The Bio21 Molecular Science and
Biotechnology Institute, The University of Melbourne, Melbourne,
VIC, Australia
2 Metabolomics Australia, School of Botany, The University of
Melbourne, Melbourne, VIC, Australia
3 Australian Bioinformatics Facility, Centre for Comparative
Genomics, Murdoch University, Murdoch, WA, Australia
1. Metabolomics Australia
Metabolomics Australia (MA) was established in 2007 and is a
research service-delivery consortium that offers high throughput
metabolomics services to life sciences researchers in academia and
industry on both an open access and research hotel basis. It was
established with funding from the Federal Government National
Collaborative Research Infrastructure Strategy (NCRIS) scheme to
Bioplatforms Australia Pty Ltd (
http://www.bioplatforms.com.au/)
and co-investment from State governments and institution partners.
The consortium is made up of five nodes at The University of
Melbourne (Victoria), Australian Wine Research Institute Ltd.
(South Australia), University of Western Australia (Western
Australia), Murdoch University (Western Australia) and University
of Queensland (Queensland). Details on the various technologies
available, analytical approaches used and the different services
provided by Metabolomics Australia are detailed at our website
http://www.metabolomics.com.au/.
MA offers both analytical services and data analysis solutions.
2. Software Solutions
The Bioinformatics Group within Metabolomics Australia, in
collaboration with the Australian Bioinformatics Facility at
Murdoch University, is developing a number of software solutions
for high throughput metabolomics data analysis and visualisation.
In this article, we detail three such solutions.
2.1 Metabolomics Australia LIMS (MASTR-MS)
Each of the Metabolomics Australia nodes handles a large
number of projects and clients/collaborators, and the development
of a flexible LIMS for capturing experimental and analytical data
has been a high priority. We have developed the MA Sample Tracking
Repository (MASTR-MS) that comprises four main modular components:
- The Node Management System controls the participation of
different laboratories that are part of the LIMS system by
allowing the option to add and delete laboratories that are
part of the LIMS.
- The User Management and Quote System provides options for
user registration including client registration, control of
different user groups and their privileges in the LIMS system.
In addition, this module allows the client the option of
interacting with laboratory staff through a quote request and
response system.
- The Sample Management System is used to capture the
metadata that describes a project and all the experiments
contained within it. Once captured, the metadata is used to
generate a sample list, which can then be run any of the
analytical instruments that are associated with the LIMS. MA
scientists and clients are able to visualise progress of their
samples through the sample tracking system.
- The Data Management System controls the transfer of raw
data from the instruments to the LIMS and its association with
relevant projects and experiments. This module also allows
laboratory users to add processed data to the LIMS system by
associating them with projects and allows clients to download
their raw and processed data.
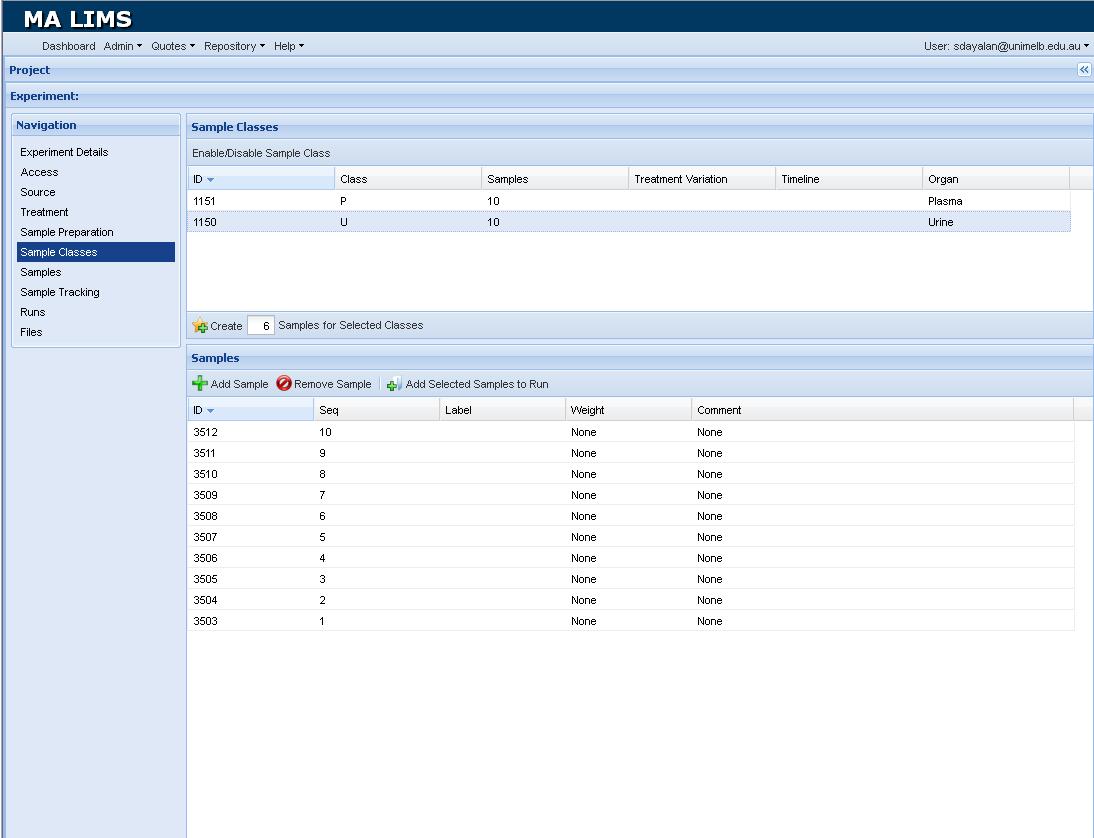
Figure 1. Screenshot of the LIMS window.
2.2 Metabolomics Australia MS
Database (MAMBO-MS)
In order to facilitate metabolite identification in GC-MS and
LC-MS analyses we have developed the
MA Meta
BOlite
Mass
Spectral database
(MAMBO-MS) as a searchable repository of GC/LC mass spectra and
associated information. MAMBO-MS was developed as an in-house
database solution, as none of the publicly available database
frameworks met MA's requirements. The requirements and the
MAMBO-MS features that were developed as a result are detailed
below. MAMBO-MS comprises four major modules:
- Lab Management System: This module allows control over the
number of laboratories/groups that contribute data to
MAMBO-MS. This module was build due to the geographically
distributed nature of MA's nodes. Through this module, more
laboratories/groups can either be added to MAMBO-MS or
existing laboratories can be deleted. Addition and deletion of
laboratories in this module would automatically reflect in the
following modules.
- User Management System: This module creates and manages
different user groups (Admin, Node Leader, Staff, Client) and
their privileges over the available features of MAMBO-MS.
- Records Management System: Controls the mass spectral
records uploaded by the different participating labs. MAMBO-MS
stores GC-MS and LC-MS records, where each record holds a
series of metadata information on the spectra acquisition
conditions (instrument, method, etc.) and information on the
mass spectra itself (source, names, molecular weight, spectral
values, etc.). In addition, MAMBO-MS stores NIST 08 records
for internal use only.
- Search System: This module allows users to search the
records by either keyword or by spectra.
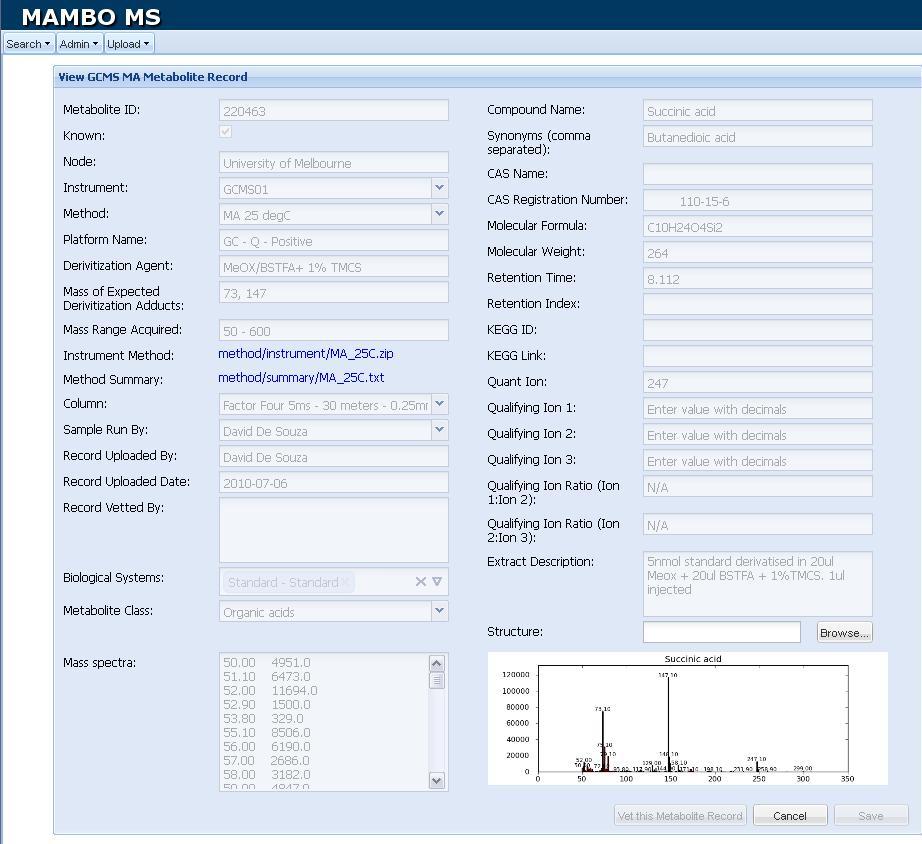 Figure 2.
Figure 2. Screenshot of a
GC-MS Record Window.
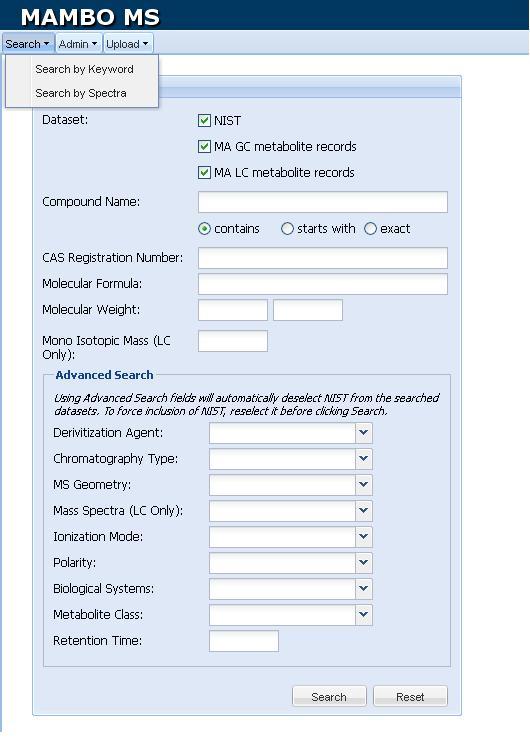 Figure 3.
Figure 3. Screenshot of
search options in MAMBO-MS Window.
2.3 PyMS
PyMS is a set of libraries for processing of GC-MS data, written
in the Python programming language. PyMS was developed to allow
high throughput alignment and peak integration of GC-MS
chromatograms generated from experiments involving hundreds of
sample runs. Together with the python programming environment,
PyMS provides a powerful framework for rapid development and
testing of methods for processing of GC-MS data [1]. Python
scripts using the PyMS framework can be quickly and easily written
for large scale data processing, providing a useful batch
processing tool. These scripts can in turn be easily incorporated
into automated data processing pipeline tools, frontends, web
servers, or any other systems capable of calling Python scripts.
Typically, a PyMS user will set up a PyMS processing pipeline in
the following way:
- For each file in the GC-MS analysis:
- Read in raw data into a PyMS Intensity Matrix object
- Filter the raw data to remove unwanted signal (noise)
- Find all peaks, and filter the peak list to remove
unrealistic peaks
- Then, for each peak list
- Align the peaks using Dynamic Programming of Peak objects
- Write out a matrix of aligned peak areas for all data
files
To calculate the area of each peak, PyMS uses a novel quantitation
method. After alignment, aligned peaks are interrogated for ions
which are both abundant and also common to each individual peak. A
single 'common ion' is chosen for each aligned peak and area
quantitation is based on this ion. Thus PyMS area quantitation is
extremely robust and the influence of noise is greatly reduced.
A set of example scripts, covering most common GC-MS processing
tasks has been created as the pyms-test project. In addition, the
pyms-docs project provides a comprehensive user manual. When
combined, pyms-test and pyms-docs provide a thorough introduction
and reference to PyMS.
2.4 Availability
All of the above detailed software projects are developed as open
source solutions under the GPL license. Their source codes are
available from:
3 Future Directions
These three software projects form different sections of the GC-MS
and LC-MS data production and processing work flow pipeline.
Future efforts are directed at integrating these software packages
into a seamless work flow system for the capture, analysis, and
visualisation of metabolomics data generated by high throughput
technologies.
[1] Seán O'Callaghan
et
al., "PyMS: a Python toolkit for processing of gas
chromatography--mass spectrometry (GC-MS) data. Application and
comparative study of selected tools",
BMC Bioinformatics, 2012, In Press.
Please
note: If you know of any
metabolomics research programs, software, databases,
statistical methods, meetings, workshops, or training
sessions that we should feature in future issues of this
newsletter, please email Ian Forsythe at metabolomics.innovation@gmail.com.











