
TMIC and the Metabolomics Society
Issue 45 - May 2015
CONTENTS:
Online version of this newsletter:
http://www.metabonews.ca/May2015/MetaboNews_May2015.htm
 |
| Published
in partnership between TMIC and the Metabolomics Society Issue 45 - May 2015 |
|
CONTENTS: |
|
|
 |
|
 |
|
 |
| Metanomics Health GmbH |
Chenomx
Inc. |
mzCloud
Mass Spectral Database |
 |
Metabolomics Society News |

 |
 |

| |
Metabolomics Spotlight
|
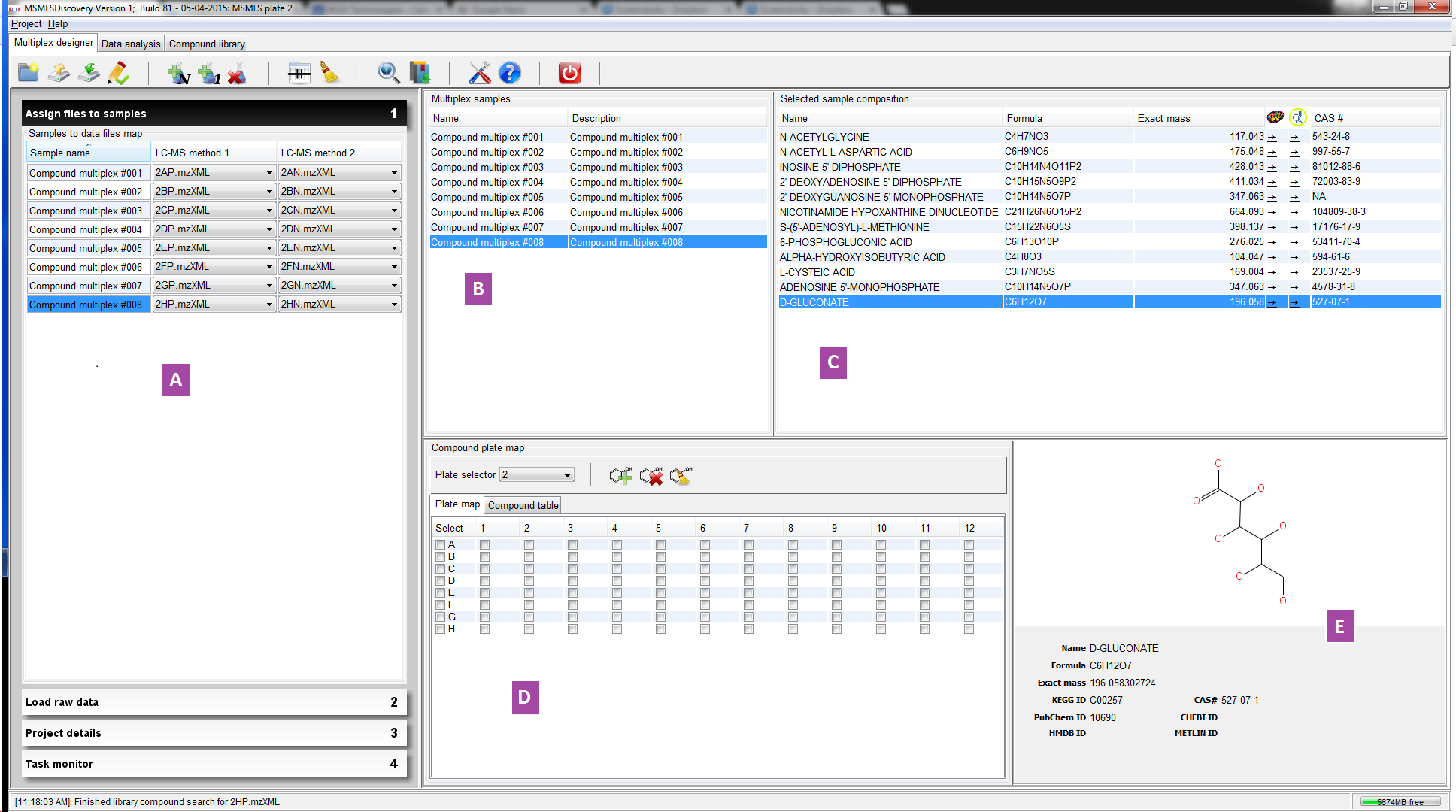
Click on the thumbnail image below to view a larger version of the image
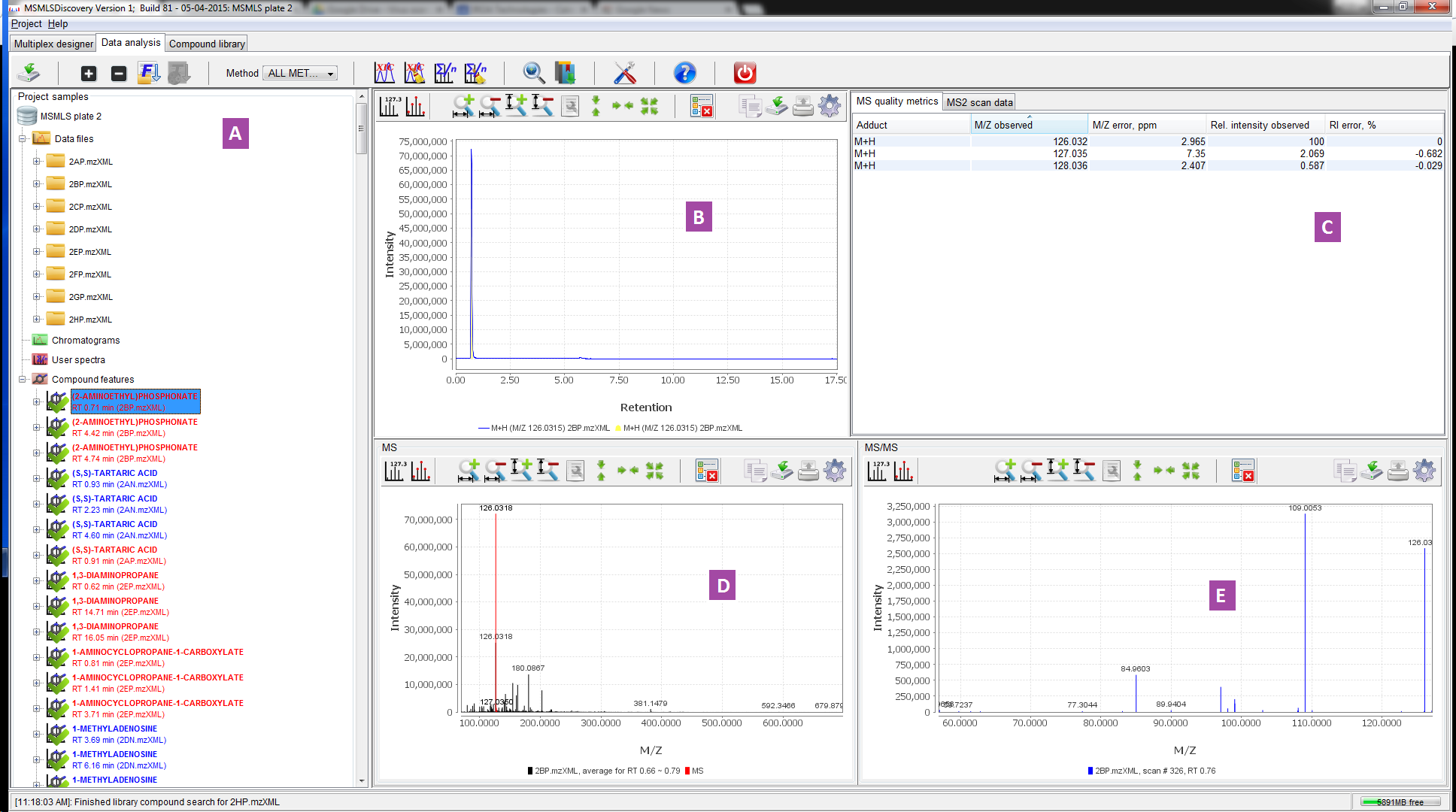
Figure 2. Data analysis panel
| |
MetaboInterview
|
| Professor
of Pharmaceutical Technologies at the University of
Greenwich, United Kingdom |
|
 |
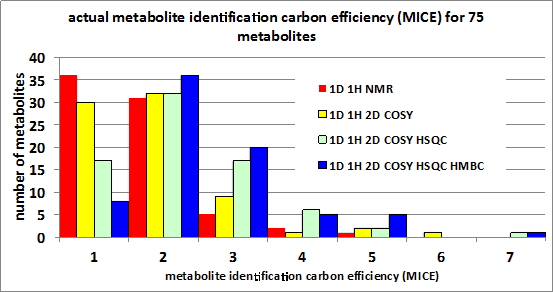
Biography Jeremy Everett is the Professor of Pharmaceutical Technologies at the University of Greenwich, UK. Jeremy previously held a variety of global drug discovery technology leadership positions for Pfizer, and before that, SmithKline Beecham. He is a consultant on lead and drug discovery to both large pharma and small research institutes, and has worked as an expert witness for a US law firm in a patent case. Jeremy conducts research in metabonomics and pharmacometabonomics, and has made several contributions in this area: he conceived and co-named the new science of metabonomics; he co-discovered and co-named pharmacometabonomics (the ability to predict the effect of a drug before dosing, based on pre-dose metabolic profiles, Nature 2006), and he also co-led the research project and clinical trial that demonstrated pharmacometabonomics in humans for the first time (PNAS 2009). He is a Visiting Professor in the Department of Surgery and Cancer at Imperial College, London. Jeremy received his BSc and PhD in chemistry from Nottingham University, UK. He did post-doctoral studies in NMR at McMaster University and at McGill University in Canada. Jeremy has published over 80 scientific papers and reviews and has one granted patent. His h-index is 24, with over 3,000 total citations. |
 |
Metabolomics Current Contents
|
This
section of MetaboNews is supported by: |
Metabolomics Events
|
| 6-7 May 2015 |
Innovations in Omics Research and
Translational Medicine Waters invites you to a free Innovations in Omics Research and Translational Medicine meeting on May 6-7th in Eschborn, Germany focused on mass spectrometry based solutions for omics research and translational medicine. Register: http://www.waters.com/omics2015 During this event you will learn about :
|
| 11-14 May 2015 |
2015 SECIM Workshop The 2nd Annual SECIM Metabolomics Workshop will be
held Monday, May 11 through Thursday, May 14, 2015, in
Gainesville, Florida. Click
here for the event flyer For more information, visit http://secim.ufl.edu/workshops/. |
| 21-22 May 2015 |
2nd Metabolomics - Advances &
Applications in Human Disease Conference Panel: |
| 14-18 Jun 2015 |
3rd Annual Workshop on Metabolomics The themes in this third year of the workshop are:
|
| 15-16 Jun 2015 |
Informatics and Statistics for
Metabolomics (2015) A poster announcing this workshop can be found here.The workshop will cover many topics ranging from understanding metabolomics technologies, data collection and analysis, using pathway databases, performing pathway analysis, conducting univariate and multivariate statistics, working with metabolomic databases and exploring chemical databases. Participants will be given various data sets and short assignments to assist with the learning process. Target Audience This course is intended for graduate students, post-doctoral fellows, clinical fellows and investigators who are interested in learning about both bioinformatic and cheminformatic tools to analyze and interpret metabolomics data. Prerequisite: Familiarity with R is required. Familiarity can be gained through online activities. You should be familiar with these R concepts (chapters 1-5) or review the past Statistics tutorials provided by CBW. Apply Now Award Opportunities For further details, visit http://bioinformatics.ca/workshops/2015/informatics-and-statistics-metabolomics-2015. |
| 15-18 Jun 2015 |
Metabolomics Summer Workshop This workshop is intended for investigators seeking a solid foundation to expand their research using metabolomics. Sessions include:
For further details, visit http://mrc2.umich.edu/Events_Summer_Workshop.php. |
| 16-19 Jun 2015 |
Metabolic Phenotyping in Disease
Diagnosis & Personalised Health Care For further details, visit http://www1.imperial.ac.uk/resources/D84E5488-E7A5-475D-8EDC-32FB0B0522A9/met_pheno_nov_2015june_preliminary.pdf. |
| 29 Jun to 2 Jul 2015 |
Metabolomics 2015: 11th Annual
Conference of the International Metabolomics Society You are invited to join us for Metabolomics 2015,
the official annual meeting of the Metabolomics
Society. For further details, visit http://metabolomics2015.org. |
| 26-28 Aug 2015 |
Bio&Data “Mountain Village
Science Series” (MOVISS 2015) For further details, visit http://www.moviss.org/. |
| 21-25 Sep 2015 |
W4M Course 2015: Computational
Metabolomics within the Galaxy Environment on the
Online Workflow4Metabolomics.org (W4M) Resource Program Schedule: http://workflow4metabolomics.org/training/W4Mcourse2015/program Practical Information: http://workflow4metabolomics.org/training/W4Mcourse2015/practical-info Preregistration (deadline of May 8, 2015): http://workflow4metabolomics.org/training/W4Mcourse2015/registration-form Organizing Committee Cécile Canlet, Christophe Caron, Franck Giacomoni, Yann Guitton, Gildas Le Corguillé, and Étienne Thévenot. For further details, visit http://workflow4metabolomics.org/training/W4Mcourse2015. |
| 7-9 Dec 2015 |
MetaboMeeting 2015 Agenda Topics
|
Metabolomics Jobs |
This is a resource for advertising positions in
metabolomics. If you have a job you would like posted in this
newsletter, please email Ian Forsythe (metabolomics.innovation@gmail.com).
Job postings will be carried for a maximum of 4 issues (8
weeks) unless the position is filled prior to that date.
Jobs Offered
| Job Title | Employer | Location | Posted | Closes | Source |
|---|---|---|---|---|---|
| Postdoc, Northwest
Metabolomics Research Center |
University of Washington | Seattle, Washington, USA | 17-Apr-2015 |
31-May-2015 or until filled |
Metabolomics
Society Jobs |
| Postdoc, Mitochondria and Metabolism Center | University of Washington | Seattle, Washington, USA | 17-Apr-2015 |
31-May-2015 or until filled |
Metabolomics
Society Jobs |
| Bioinformatician (Metabolomics) |
Leibniz Institute of Plant Biochemistry | Halle, Germany | 8-Apr-2015 |
Open until filled |
Leibniz
Institute of Plant Biochemistry |
| Research Assistant I, Metabolomics Core
Facility |
Sanford-Burnham
Medical Research Institute |
Orlando, Florida, USA | 3-Mar-2015 |
Open until filled |
Metabolomics
Society Jobs |
| Postdoctoral Research Fellow, Fernandez
Laboratory, School of Chemistry and Biochemistry |
Georgia Institute of Technology |
Atlanta, Georgia, USA | 19-Feb-2015 |
Open until filled | Georgia Institute of Technology |
| Computational Metabolomics Professor |
Pennsylvania State University |
State College, Pennsylvania, USA |
26-Jan-2015 |
Metabolomics Society Jobs |
|
Ian J. Forsythe Double AB, MSc Editor, MetaboNews Department of
Computing Science
University of Alberta 221 Athabasca Hall Edmonton, AB, T6G 2E8, Canada Email: metabolomics.innovation@gmail.com Website: http://www.metabonews.ca LinkedIn: http://ca.linkedin.com/in/iforsythe Twitter: http://twitter.com/MetaboNews Google+: https://plus.google.com/118323357793551595134 Facebook: http://www.facebook.com/metabonews |
This newsletter is published in
partnership between The Metabolomics Innovation Centre
(TMIC, http://www.metabolomicscentre.ca/) and the Metabolomics
Society (http://www.metabolomicssociety.org)
for the benefit of the worldwide metabolomics
community.
|