
TMIC and the Metabolomics Society
Issue 57 - May 2016
CONTENTS:
Online version of this newsletter:
http://www.metabonews.ca/May2016/MetaboNews_May2016.htm

 |
| Published
in partnership between TMIC and the Metabolomics Society Issue 57 - May 2016 |
|
CONTENTS: |
|
|
 |
| TMIC
Services |
 |
Metabolomics Society News |
| |
Metabolomics Spotlight
|
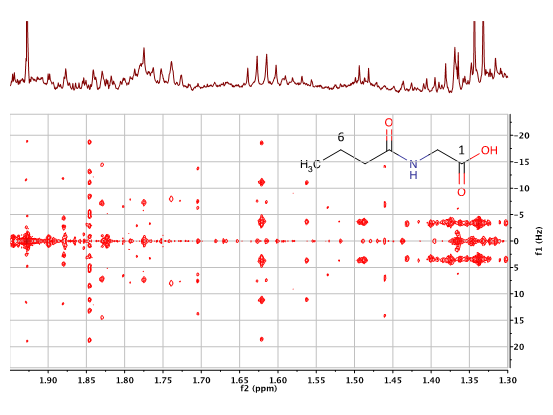
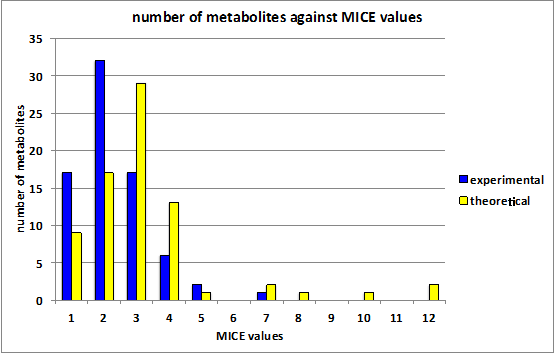
| |
MetaboInterview
|
| Director,
Systems and Translational Sciences, RTI International,
Research Triangle Park, NC, USA |
|
 |
Biography Susan Sumner is working to make personalized medicine a reality. Using metabolomics, the unique chemical fingerprints that cellular processes leave behind, Sumner assesses differences in the metabotype of individuals that correlate with states of wellness or disease. She is identifying individuals' response to treatment in areas such as obesity, drug-induced liver injury, infectious disease, and reproductive and developmental biology. She is also identifying biomarkers for the early detection and diagnosis of disease to monitor therapeutic treatments and to provide insights into biological mechanisms. Dr. Sumner is the director of the Systems and Translational Sciences Center in the Discovery Sciences Division at RTI, where she oversees the proteomics and metabolomics cores. In September 2012, Dr. Sumner and her team established the NIH Eastern Regional Comprehensive Metabolomics Resource Core (RTI RCMRC) through a grant from the National Institutes of Health (NIH) Common Fund. The RTI RCMRC works with an NIH-funded consortium to establish nationwide standards for metabolomics data collection and analysis, data reporting and deposition, and to provide training to basic and translational researchers. Dr. Sumner serves on the editorial boards for Environmental Health Perspectives, Metabolomics, the Journal of Applied Toxicology, and the Journal of Toxicology, and is an adjunct faculty member in the Brody School of Medicine at East Carolina University, and in the Department of Nutrition at the University of North Carolina at Chapel Hill. http://www.rti.org/newsroom/experts.cfm?obj=1958B5A7-73A2-4C7B-B3502AB6EB5F5AB3 |
 |
Metabolomics Current Contents
|
Metabolomics Events
|
| 3-4 May 2016 |
2016 SECIM Metabolomics Symposium The two-day symposium will review the basics of metabolomics, while showcasing the application of metabolomics in a variety of fields. Registration is FREE! The event is appropriate for graduate students, technicians, postdocs and faculty new to the field. RSVP Deadline: Friday, April 22 Please feel free to distribute the 2016 SECIM Metabolomics Symposium flyer to any interested persons. For more information and to register for this event, visit http://secim.ufl.edu/education/events/. |
| 9-12 May 2016 |
Metabolomics Summer Workshop Registration: https://umichumhs.ut1.qualtrics.com/SE/?SID=SV_bPEVDn7I5wSEcwl This workshop is intended for investigators seeking a solid foundation to expand their research using metabolomics. Sessions include:
If you have any questions about the workshop, please contact Terri Ridenour. For more information, visit the MRC2 website. |
| 9 May-3 June 2016 |
Metabolomics: Understanding
metabolism in the 21st Century Study time: 4 weeks, 3 hours study time per week. You will have access to the course after the end date. The course is aimed at final year undergraduate science students and research scientists but will provide a valuable introduction to the metabolomics field for scientists at any stage in their careers. To register for this free online course please visit https://www.futurelearn.com/courses/metabolomics |
| 26-27 May 2016 |
Informatics and Statistics for
Metabolomics (2016) The workshop will cover many topics ranging from understanding metabolomics technologies, data collection and analysis, using pathway databases, performing pathway analysis, conducting univariate and multivariate statistics, working with metabolomic databases and exploring chemical databases. Participants will be given various data sets and short assignments to assist with the learning process. For more information and to register, visit http://bioinformatics.ca/workshops/2016/informatics-and-statistics-metabolomics-2016. |
| 30 May-2 Jun 2016 |
10th conference of the
Francophone Metabolomics and Fluxomics Society
(RFMF)
|
| 14-17 Jun 2016 |
Metabolic Phenotyping in Disease
Diagnosis and Personalised Health Care Lectures will cover data acquisition and analysis with some advanced statistical workshops for more hands-on participation for attendees. There will also be examples of real life applications from the research at Imperial College and their collaborators. Objectives include:
http://www.imperial.ac.uk/imperial-international-phenome-training-centre/courses/metabolic-phenotyping-in-disease-diagnosis-and-personalised-health-care or contact Dr Liz Want (iptc@imperial.ac.uk) for further information. |
| 21-24 Jun 2016 |
Data Analysis for Metabolic
Phenotyping It combines lectures and tutorial sessions to ensure a thorough understanding of the theory and practical applications.
http://www.imperial.ac.uk/imperial-international-phenome-training-centre/courses/hands-on-data-analysis-for-metabolic-profiling or contact Dr Tim Ebbels (iptc@imperial.ac.uk) for further information. |
| 27-30 Jun 2016 |
Metabolomics 2016 The conference programme will cover all aspects of metabolomics under five broad themes: Metabolomics in Nutrition and Food Research, Metabolomics in Health and Disease, Advancing the Field, Environmental, Plant and Model Organisms. For further details, visit http://metabolomics2016.org. |
| 17-21 Jul 2016 |
4th Annual Workshop on
Metabolomics Women, members of under-represented minority groups, and individuals with disabilities are strongly encouraged to apply. The metabolome, in distinction to the other –Omics, provides a better assessment of the exposure to the chemicals of life – it is truly intergenomic providing information on “metabolites” coming from ourselves or the model we’re using, the unique, non-human/mammalian compounds we consume as part of our food, compounds generated by the microorganisms that are part of “us”, and other compounds that are in the environments we live in. If ever precision medicine is able to deliver what is claimed it will, it has to include the metabolome context in which genes and proteins are operating. The themes in this fourth year of the workshop are:
|
| 12-23 Sep 2016 |
2016 International Summer
Sessions in Metabolomics
|
Metabolomics Jobs |
This is a resource for advertising positions in
metabolomics. If you have a job you would like posted in
this newsletter, please email Ian Forsythe (metabolomics.innovation@gmail.com).
Job postings will be carried for a maximum of four issues
(eight weeks) unless the position is filled prior to that
date.
Jobs Offered
| Job Title | Employer | Location | Posted | Closes | Source |
|---|---|---|---|---|---|
| Postdoctoral Associate - Biochemistry |
University of Florida |
Main Campus, Gainesville, FL, USA | 2-May-2016 | 2-Jun-2016 |
University
of Florida |
| Postdoctoral Associate Position in
Metabolomics |
Yale University | New Haven, USA | 28-Apr-2016 | |
Yale
University |
| Field Application Specialist
Metabolomics/Mass spectrometry (East Coast) |
BIOCRATES Life Sciences |
East Coast, USA | 26-Apr-2016 | |
BIOCRATES
Life Sciences AG |
| Metabolomics Method Development
Specialist |
Purdue University |
West Lafayette, Indiana, USA | 19-Apr-2016 | |
Metabolomics
Society Jobs |
| Postdoctoral Scientist |
University of Alberta |
Edmonton, Canada | 13-Apr-2016 | |
The
Metabolomics Innovation Centre |
| Research Postdoctoral Scientist |
Beaumont Health System | Michigan, USA | 31-Mar-2016 | |
Beaumont
Health System |
| Postdoctoral Position in Tumor
Metabolism |
Michigan State University | East Lansing, MI USA | 16-Mar-2016 | Until filled | Metabolomics Society |
| Assistant or Associate Professor |
University of Nebraska-Lincoln |
Lincoln, NE, USA | 4-Mar-2016 | Until filled | Metabolomics Society |
|
Ian J. Forsythe Double AB, MSc Editor, MetaboNews Department of
Computing Science
University of Alberta 221 Athabasca Hall Edmonton, AB, T6G 2E8, Canada Email: metabolomics.innovation@gmail.com Website: http://www.metabonews.ca LinkedIn: https://www.linkedin.com/in/ijforsythe Twitter: http://twitter.com/MetaboNews Google+: https://plus.google.com/118323357793551595134 Facebook: http://www.facebook.com/metabonews |
This newsletter is published in
partnership between The Metabolomics Innovation
Centre (TMIC, http://www.metabolomicscentre.ca/) and the Metabolomics
Society (http://www.metabolomicssociety.org)
for the benefit of the worldwide metabolomics
community.
|