IROA™: An Accurate Quantitative Biochemical Profiling Tool
Feature article contributed by Felice de Jong,
CEO, NextGen Metabolomics, Inc., Ann Arbor, Michigan, USA
IROA™ (Isotopic Ratio Outlier Analysis) is a semi-automated
metabolic profiling tool developed by NextGen Metabolomics, Inc. (
www.nextgenmetabolomics.com)
to assist the researcher in the identification and quantitation of
biochemical metabolites. Current mass spectral techniques used in
metabolomics make it difficult to identify and characterize the
peaks, and differentiate valid biological peaks from artifacts.
Often the researcher cannot easily obtain enough information to
accurately identify and name compounds, and less abundant spectral
signals get buried in the noise and discarded. The IROA protocol
and software side-step many of these problems making it easy to
remove confounding artifacts and noise from the dataset and
readily calculate the number of carbons for each identified
metabolite in order to verify the identity of each molecule.
Metabolite identification and quantitation is based on creating a
full complement of internal standards for the metabolome under
study, providing triply redundant information and maximizing
quality control in the process. Users can perform metabolomic
analyses with either
in vitro generated experimental
samples or with samples collected from bodily fluids or tissues.
The basic principle of IROA
IROA is an unbiased biochemical profiling protocol that relies on
differential labeling of the carbon-backbone of two groups of
samples. This is achieved by growing cells in media made
specifically for the IROA protocol in which all carbon sources
have been appropriately labeled so they are chemically identical,
but isotopically different. Briefly, a homogenous cell population
is divided into two groups, "control" and "experimental", and each
group is labeled with a distinct specific isotopic ratio, namely
95%
13C: 5%
12C
and 5%
13C: 95%
12C,
respectively (
Figure
1). After sufficient growth, at least five cell divisions,
the original carbon content of each of these cell populations is
fully diluted and replaced with the isotopic balance of the media.
In an experiment, the experimental population is treated with a
stressor, and the control population is treated with vehicle.
After the experiment is complete, a single experimental sample and
a control sample are combined, prepped as a composite sample, and
analyzed using LC-MS. Software algorithms written for the IROA
protocol are employed to identify all the different isotopic
patterns, sort the biological signals, remove artifacts,
normalize, and quantitate the relative ratios of experimental to
control analytes in each sample. The software reduces the
complexity of the entire mass spectrum into a list of biochemicals
identified in the sample and reports how each metabolite deviates
from its common internal control.
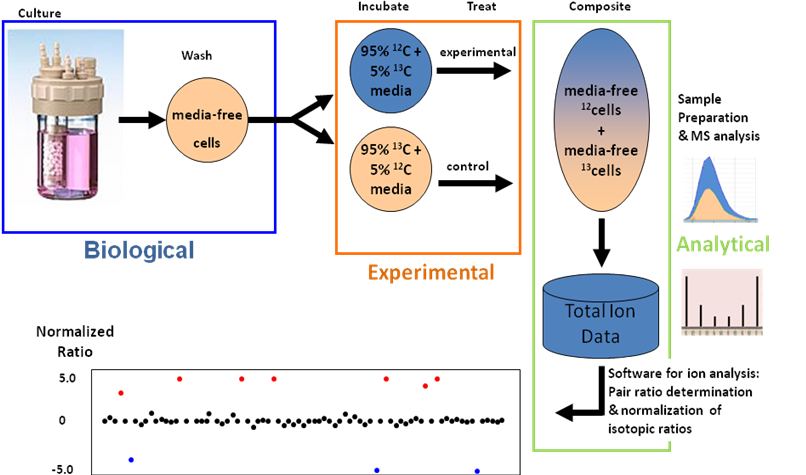 Figure 1. IROA Method for determining the biological response
to drugs, toxins, or other stressors.
Figure 1. IROA Method for determining the biological response
to drugs, toxins, or other stressors. IROA isotope ratios
of
12C/
13C
95%/5% and 5%/95% allow for control and experimental samples to be
run simultaneously, eliminating sample-to-sample variability.
Highlights of the IROA protocol
The isotopic signature created in all biological molecules during
the IROA labeling process imparts additional analytical advantages
compared with standard isotopic labeling. One of the biggest
advantages is that when using the IROA protocol, since both groups
of biological molecules (control and experimental) are labeled
with distinct biochemical signatures, the origin of every peak in
the composite sample can be readily determined and artifacts and
noise can be easily distinguished as they contain only natural
abundance carbon.
IROA metabolites derived from control and experimental samples
have enhanced M-1 and M+1 peaks, respectively, which can be used
to calculate the number of carbons in each analyte (
Figure
2). The M-1 and M+1 peak heights relative to the base peak
of each molecule can be effectively calculated by the binomial
expansion of the expression: (12C% +13C%)
N
where N equals the number of carbons, and
12C%
and
13C% equals the relative isotopic
abundances. This is not sufficiently accurate when all atoms are
present at their natural abundance, but at 5% and 95%, the M+1 and
M-1 peaks are diagnostic and indicate where the paired mate will
be located. The number of carbons in a biological molecule can be
determined by the distance between the two base peaks,
12C
and
13C, and the relative height of the
M+1 and M-1 provide confirmation of the fact, providing triply
redundant information. The number of carbons in a molecule
together with the accurate mass of the C
12
and C
13 monoisotopic peaks constrain the
number of potential formulae so that the identification of the
peaks for almost all metabolites is accurately made to the unique
formula.
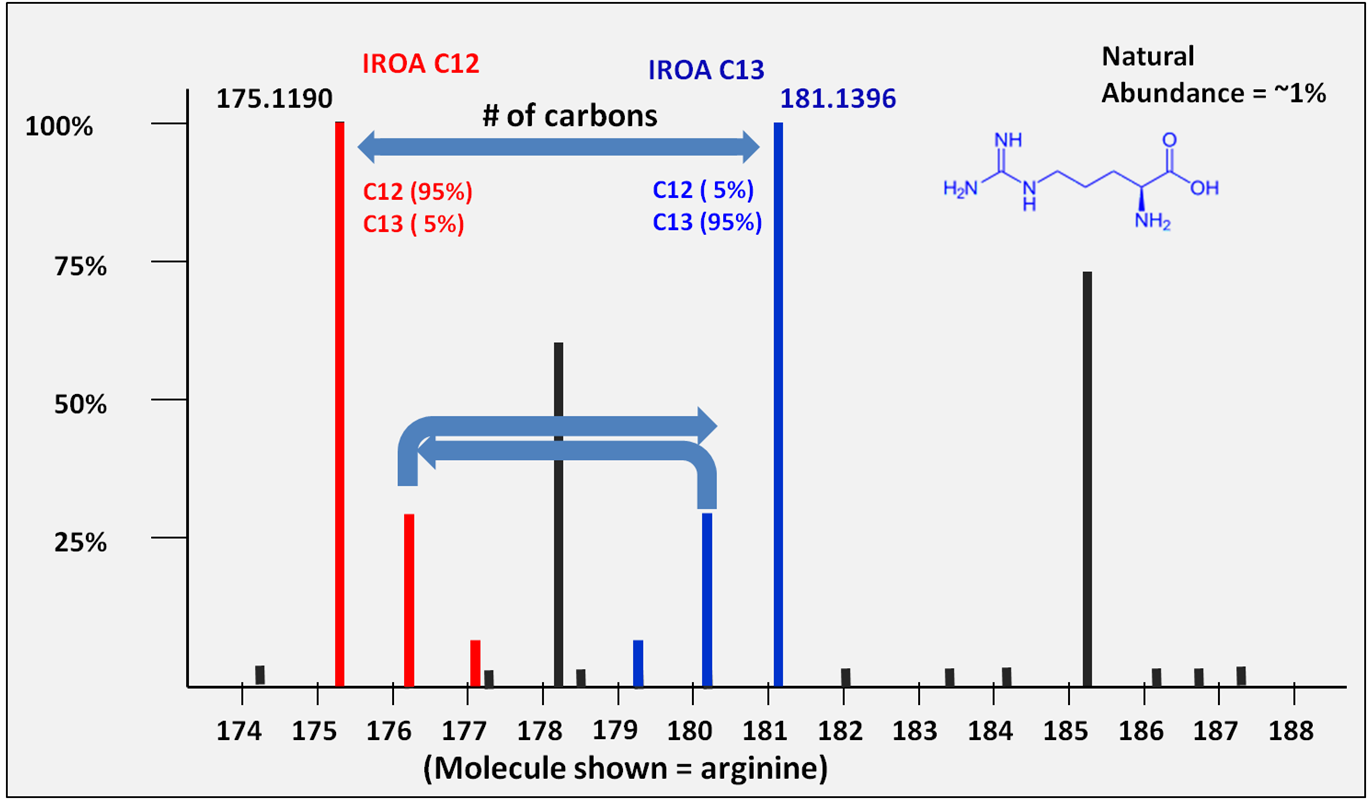 Figure 2. The IROA Peaks.
Figure 2. The IROA Peaks. In the case of arginine, the
12C
M
+ located at 175.1190 and its
13C
mate at 181.1396 clearly indicate a 6 carbon molecule. The
corresponding M
+1 and M
-1
peaks are a mass difference of 1.00335 amu (the mass difference
between a
13C and
12C
isotope). Natural abundance peaks from exogenous sources do not
have a
13C counterpart and are not
considered in the analysis.
Similar to other protocols including SILAC, the IROA control
sample is embedded into the IROA experimental sample prior to
sample preparation and analysis, thereby removing sample-to-sample
variance and also reducing the total number of samples to be
analyzed. Perhaps the most important of the sample-to-sample
variances controlled for by IROA, is ion suppression. This happens
because the control and experimental, analyzed as a single sample,
assure that the control and experimental quantification for every
compound is measured simultaneously.
The IROA Phenotyping approach for bodily fluids and tissues
Where it is not possible to isotopically label experimental
samples, the IROA Phenotyping protocol can be applied (
Figure
3). Natural carbon abundance experimental samples are mixed
with a fully predefined IROA "Standard" that has been isotopically
labeled at 95%
13C. Although they do
not carry any isotopic label, the exact mass and position of
compounds in the experimental samples are established relative to
the fully-defined Standard. Artifacts and noise can be identified
as they have no match. Whereas in a basic IROA dataset the ratio
of the peak areas represents the relative deviation of the
metabolic pool sizes brought about by the experimental condition,
in a Phenotyping experiment the overall pattern of deviations from
the standard will define phenotype by difference from the
Standard. A Phenotyping experiment is considered a complex
targeted analysis relative to the unbiased analysis of the full
basic IROA experiment. An ideal Standard is one that represents
the entire metabolome of the fluid or tissue under study and could
be generated by the use of IROA grown cell lines (the IROA
Standard) to achieve accurate quantitation.
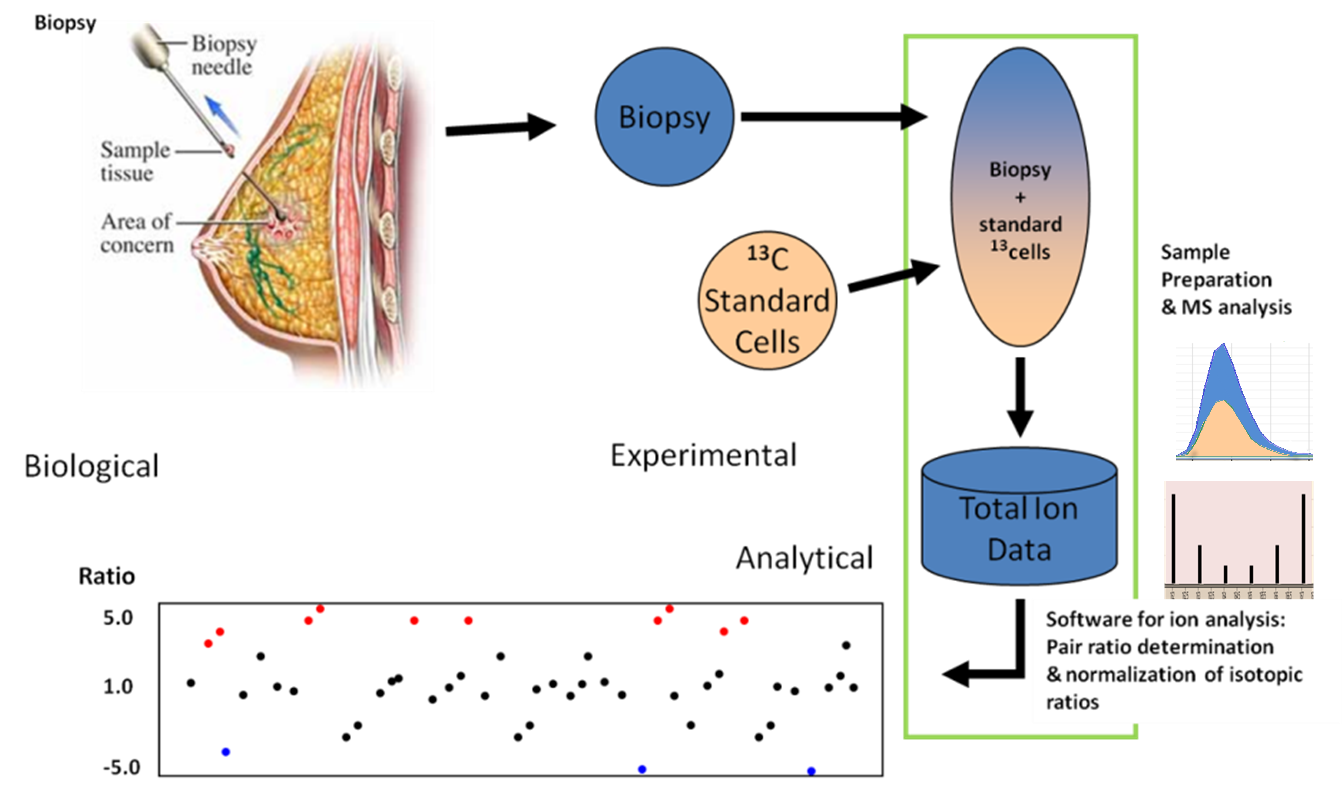 Figure 3. The IROA Phenotyping Application.
Figure 3. The IROA Phenotyping Application. The material
to be phenotyped is mixed with
13C
(IROA) cells and/or standard compounds which allow one to find and
pair all peaks. The deviation from the standard is diagnostic of
the sample's biochemical phenotype.
IROA data processing and output
The IROA peaks are all mathematically calculable and each group (
12C
and
13C) of carbon isotopomers will
reliably and accurately account for the other group providing a
redundant quality control checkpoint. The IROA software algorithms
written in Java (ClusterFinder
™) achieve a
data reduction of complex raw data to concise, high value
information, in the following steps:
- Characterization of all peaks according to source; artifact,
experimental (12C), control (13C),
or Standard
- Removal of all artifacts
- Alignment and pairing of all remaining peaks across all
scans
- Normalization and identification of all pairs
- Review/curation of the entire data set
- Determination of the relative 12C/13C
ratios of analytes in each sample
- Determination of the statistical variance of the sample
ratios
Experimental compounds that have a ratio that is a significant
deviation (greater than two standard deviations) from the average
ratio of the control group have been affected by the experimental
stressor.
Figure
4 shows representative IROA peaks for the compound
glutathione from the application of IROA in a biological system,
Saccharomyces
Cerevisiae S288C, grown in an aerobic culture during a
72-hour time course.
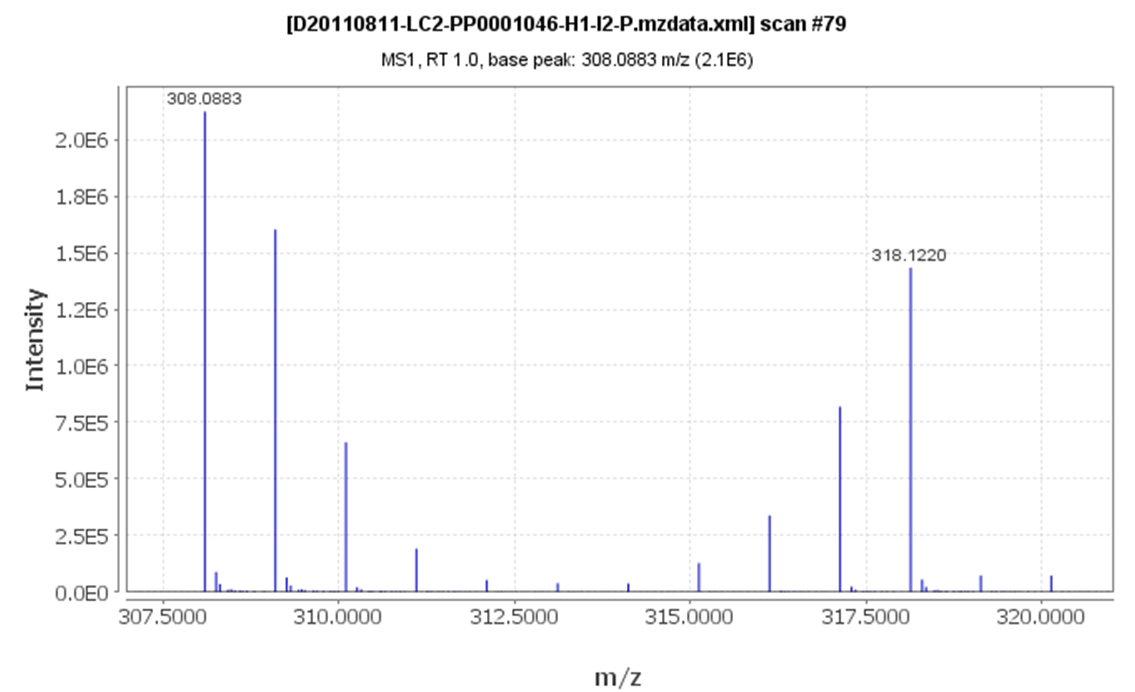 Figure 4. A Characteristic IROA Compound.
Figure 4. A Characteristic IROA Compound. Glutathione
exhibits characteristic IROA peaks; namely enhanced M+1, M+2, etc.
that are paired with enhanced M-1, M-2, etc. peaks. At the 48 hour
time point shown here, glutathione levels have increased relative
to the 24 hour control.
Availability and future directions
NextGen Metabolomics, Inc. is working with collaborators
interested in testing the IROA technology to address a biochemical
profiling/metabolomics problem and welcomes inquiries.
ClusterFinder and IROA media are expected to be available for
researchers in 2013.
Please
note: If you know of any
metabolomics research programs, software, databases,
statistical methods, meetings, workshops, or training
sessions that we should feature in future issues of this
newsletter, please email Ian Forsythe at metabolomics.innovation@gmail.com.












