
TMIC and the Metabolomics Society
Issue 51 - November 2015
CONTENTS:
Online version of this newsletter:
http://www.metabonews.ca/Nov2015/MetaboNews_Nov2015.htm

 |
| Published
in partnership between TMIC and the Metabolomics Society Issue 51 - November 2015 |
|
CONTENTS: |
|
|
 |
| TMIC
Services |
 |
Metabolomics Society News |
For the scientific session proposals:
Title:
Content: (expected content in bullet point form)
Potential Keynote speaker:
Rationale: (can incorporate the bullet points listed above also).
For workshop proposals:
Title:
Topic Outline/Content (i.e. content in bullets):
Target Audience:
Potential Speakers:
Rationale:

| |
Metabolomics Spotlight
|
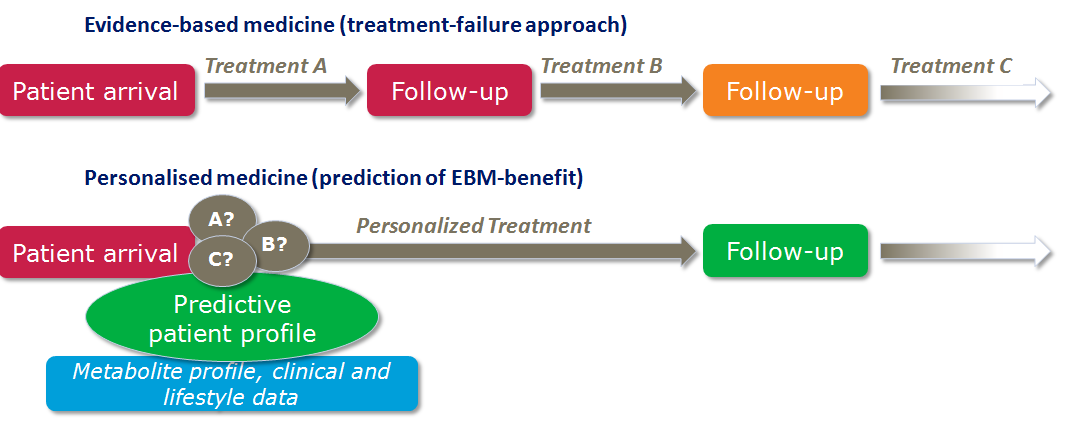
| |
MetaboInterview
|
| Senior
Scientist, RIKILT Wageningen University & Research
Centre, Wageningen, Netherlands |
|
 |
Biography Arjen Lommen is a senior scientist at RIKILT Wageningen UR. RIKILT is a government-oriented institute mainly doing research in food safety (http://www.wageningenur.nl/en/Expertise-Services/Research-Institutes/rikilt.htm). Starting out by studying protein biochemistry, molecular genetics, and technical microbiology (Molecular Sciences, 1985, WUR), he got his PhD in NMR studies on a bacterial protein. During a brief postdoc period at the Dutch National NMR facilities in 1990 (3D NMR on savinase), he taught himself programming in C. In 1991, he accepted his current job at RIKILT (part of Wageningen University and Research Center) and has gradually moved from metabolic profiling using NMR to researching and programming data analysis for high-end MS technology. During his 25 years at RIKILT, he authored and co-authored many papers in high-ranking journals in data analysis as well as applications. He is the author of a number of software packages and programs, amongst others metAlign, metAlignID, Search_LCMS, Search_GCMS, HR3—see also http://www.metabonews.ca/Dec2012/MetaboNews_Dec2012.htm. |
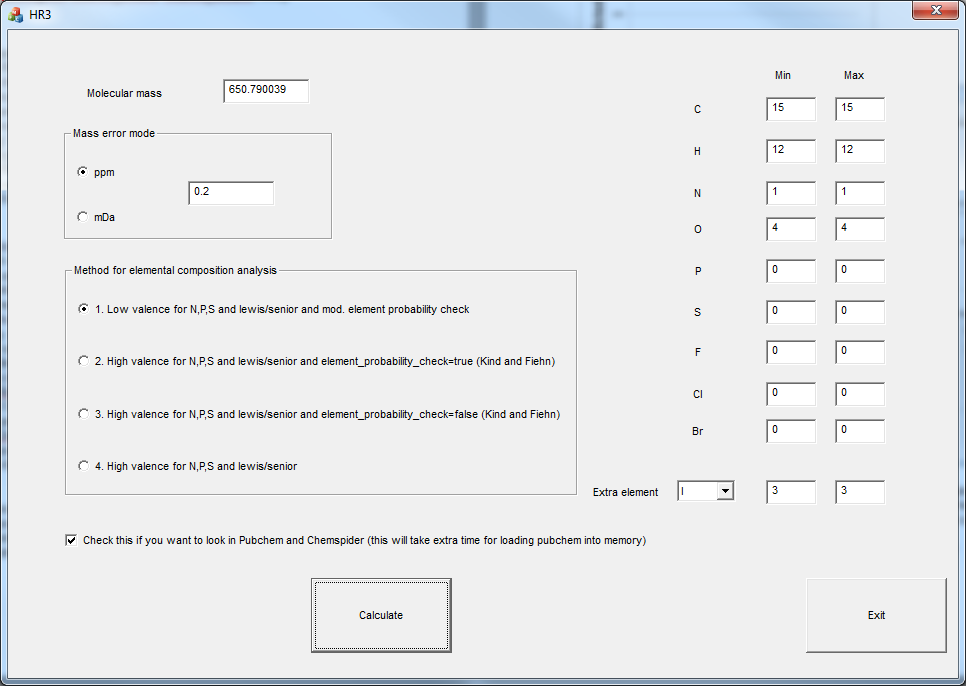
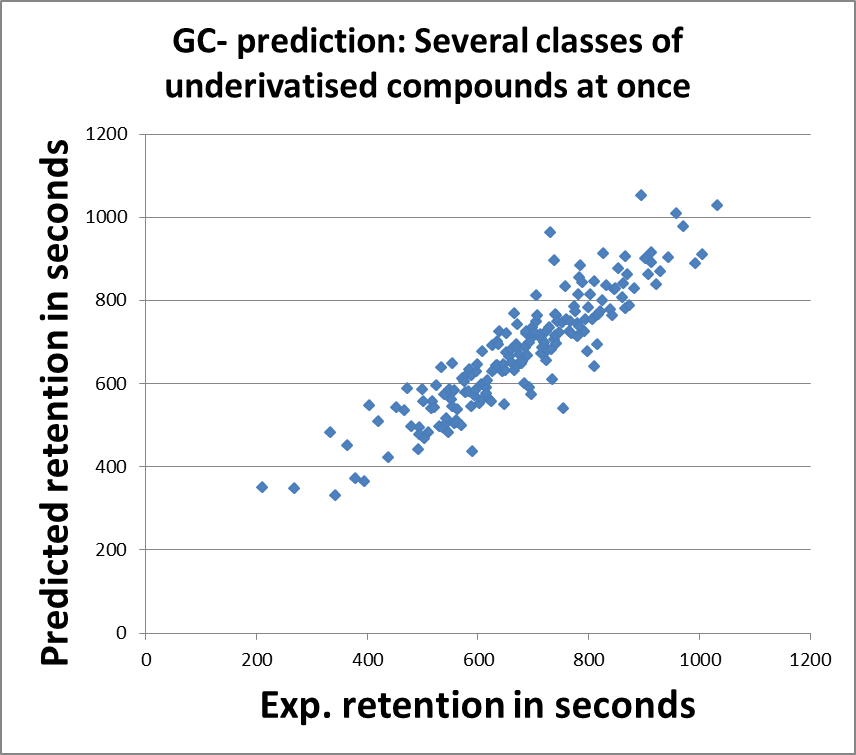
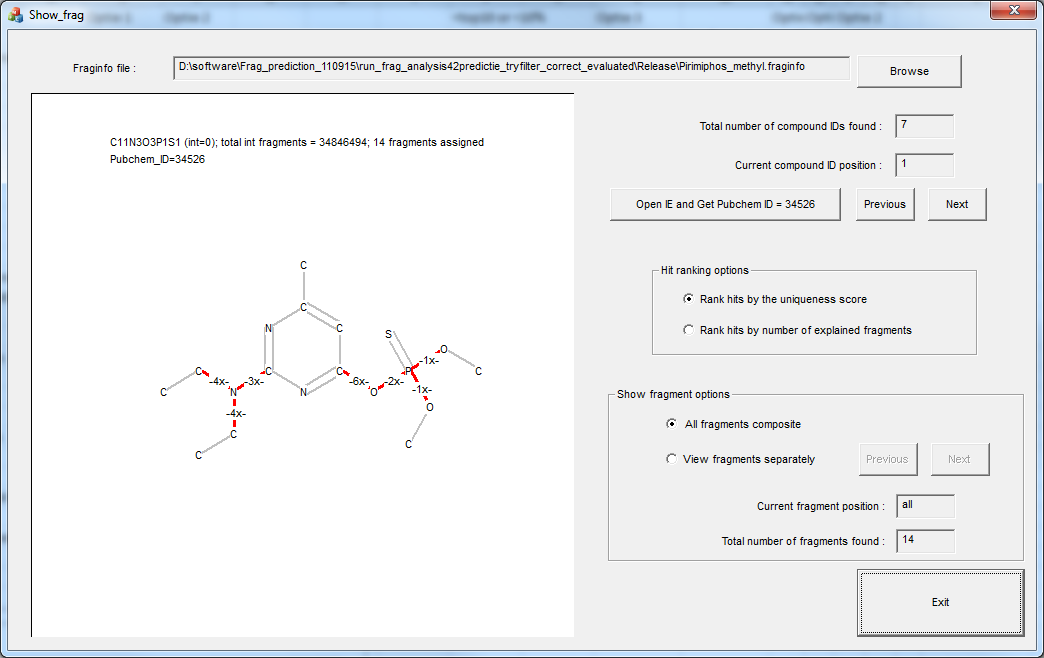
 |
Metabolomics Current Contents
|
Metabolomics Events
|
| 3-6 Nov 2015 |
Data Analysis for Metabolic
Phenotyping It combines lectures and tutorial sessions to ensure a thorough understanding of the theory and practical applications.
Please visit our website at http://www1.imperial.ac.uk/iiptc/ or contact Dr Tim Ebbels (iptc@imperial.ac.uk) for further information. |
| 7-9 Dec 2015 |
MetaboMeeting 2015 Agenda Topics
|
| 10-12 Dec 2015 |
3rd ICAN Conference Series on Omics ICAN Conference Series is an educational organization directed and managed by ICAN and the scientific community. Since its inception in 2012, ICAN organized two ICAN conference in 2013 and 2014, and they both were a real success! ICAN conference Series convene dynamic, open, peer-reviewed conferences on the exciting new frontiers of life science. Whether you are a geneticist, an immunologist or virtually any other type of life science investigator, and whether you are from academia, industry or the government/non-profit sector, we think you will find the experience of attending a ICAN Conference Series meeting valuable and memorable. As in the previous editions, we aim to warrant high quality scientific programme and facilitate interactions especially between speakers and young investigators and researchers. To that aim ICAN CONFERENCE SERIES scientific board will select young researchers abstracts eligible for Junior researchers registration rate and limit the total number of participants. Moreover, this year, certain young researchers will have the opportunity to be one of the two chairs of each session. For further details, visit http://www.ican-series.com/home. |
| 11-12 Feb 2016 |
Metabolite identification with the
Q Exactive and LTQ Orbitrap The course will be taught by experts in the fields of metabolomics and chemical analysis and will include significant hands-on experience using both the Q Exactive and LTQ-Orbitrap mass spectrometers and related software/databases/libraries. The course will focus on:
For further information and registration details, please visit http://www.birmingham.ac.uk/facilities/metabolomics-training-centre/courses/metabolite-identification.aspx or contact Dr Cate Winder (C.L.Winder@bham.ac.uk). |
| 14-19 Feb 2016 |
EMBO Practical Course on
Metabolomics Bioinformatics for Life Scientists We will have lively and interactive discussion throughout the course in addition to the hands-on data analysis and processing. We encourage you to bring your data, problems you might have with a particular data set or study for group discussion. You will be asked to present your work and participate in the discussions from day one. Who is this course aimed at: This course is aimed at PhD students, post-docs and researchers with at least two or three year’s experience in the field of metabolomics who are seeking to improve their skills in metabolomics data analysis. Topics covered:
|
| 23-24 Feb 2016 |
Quality Assurance and Quality
Control in Metabolic Phenotyping
For further information and registration details, please visit http://www.birmingham.ac.uk/facilities/metabolomics-training-centre/courses/quality-phenotyping.aspx or contact Dr Cate Winder (C.L.Winder@bham.ac.uk). |
| 30 Mar to 1 Apr 2016 |
The Australian & New Zealand
Metabolomics Conference (ANZMET 2016) Its aim is to promote collaboration, innovation, and community-building in metabolomics across private and government sectors by open engagement of all researchers and institutions throughout Australia and New Zealand. The conference is actively supported by the Australia & New Zealand Metabolomics Network (ANZMN), in conjunction with the Metabolomics Society and Metabolomics Australia. Several keynote speakers will present at the conference, including Professor David Wishart amongst others.
|
| 4-6 Apr 2016 |
Metabolomics with the Q Exactive
For further information and registration details, please visit http://www.birmingham.ac.uk/facilities/metabolomics-training-centre/courses/q-exactive.aspx or contact Dr Cate Winder (C.L.Winder@bham.ac.uk). |
| 18 Apr to 26 May 2016 |
Copenhagen School of Chemometrics -
2016 (CSC-2016)
How to enroll in the CSC-2016? Please, send an e-mail to Jose Amigo (jmar@food.ku.dk) with the information requested in this flyer. |
| 20-22 Apr 2016 |
Multiple biofluid and tissue types,
from sample preparation to analysis strategies for
metabolomics
|
Metabolomics Jobs |
This is a resource for advertising positions in
metabolomics. If you have a job you would like posted in this
newsletter, please email Ian Forsythe (metabolomics.innovation@gmail.com).
Job postings will be carried for a maximum of four issues
(eight weeks) unless the position is filled prior to that
date.
Jobs Offered
| Job Title | Employer | Location | Posted | Closes | Source |
|---|---|---|---|---|---|
| Field Application Specialist
Metabolomics/Mass spectrometry |
Biocrates Life Sciences |
East Coast, USA |
21-Oct-2015 |
|
Biocrates Life Sciences |
| An academic position in Metabolomics (1
EFT) |
Université Catholique de Louvain | Louvain-la-Neuve, Belgium |
19-Oct-2015 |
|
Université Catholique de Louvain |
| Bioinformatics (Metabolomics-specific)
Research Fellow 1 |
University of Birmingham |
Birmingham, UK | 15-Oct-2015 |
|
University of Birmingham |
| Scientist, Section of Nutrition and
Metabolism/Biomarkers Group |
International Agency for Research on Cancer | Lyon, France |
9-Oct-2015 |
4-Nov-2015 | International Agency for Research on Cancer |
| Postdoctoral research position in chemical
ecology / (imaging) mass spectrometry |
Max Planck Institute for Chemical Ecology |
Jena, Germany | 2-Oct-2015 |
|
Max Planck Institute for Chemical Ecology |
| Four Tenure-Track Faculty Positions in the
School of Integrative Plant Science |
Cornell University | Ithaca, NY, USA | 14-Sep-2015 |
Until filled | Metabolomics Society Jobs |
| Assistant/Associate Professor in Plant
Metabolomics |
Cornell University | Ithaca, NY, USA | 11-Sep-2015 |
Until filled | Metabolomics Society Jobs |
| Postdoctoral Position in Metabolomics
Bioinformatics |
University of North Carolina | Charlotte, NC, USA | 9-Sep-2015 |
Until filled | Metabolomics Society Jobs |
|
Ian J. Forsythe Double AB, MSc Editor, MetaboNews Department of
Computing Science
University of Alberta 221 Athabasca Hall Edmonton, AB, T6G 2E8, Canada Email: metabolomics.innovation@gmail.com Website: http://www.metabonews.ca LinkedIn: http://ca.linkedin.com/in/iforsythe Twitter: http://twitter.com/MetaboNews Google+: https://plus.google.com/118323357793551595134 Facebook: http://www.facebook.com/metabonews |
This newsletter is published in
partnership between The Metabolomics Innovation Centre
(TMIC, http://www.metabolomicscentre.ca/) and the Metabolomics
Society (http://www.metabolomicssociety.org)
for the benefit of the worldwide metabolomics
community.
|