
MetaboNews
Issue 3 - October 2011
CONTENTS:
Online version of this newsletter:
http://www.metabonews.ca/Oct2011/MetaboNews_Oct2011.htm
 |
| MetaboNews Issue 3 - October 2011 |
|
CONTENTS: |
|
|
Welcome to the third issue of
MetaboNews, a monthly newsletter for the worldwide
metabolomics community. In this month's issue, we feature a Software
Spotlight article on two different approaches to the
interpretation of metabolomic data through pathway
analysis and visualization using two software packages, MetScape
and MetPA. This
newsletter is being produced by The Metabolomics Innovation
Centre (TMIC, http://www.metabolomicscentre.ca/) and is
intended to keep metabolomics researchers and other
professionals informed about new technologies, software,
databases, events, job postings, conferences, training
opportunities, interviews, publications, awards, and other
newsworthy items concerning metabolomics. We hope to provide
enough useful content to keep you interested and informed and
appreciate your feedback on how we can make this newsletter
better (metabolomics.innovation@gmail.com).
Note: Our subscriber list
is managed using Mailman, the GNU Mailing List Manager. To
subscribe or unsubscribe, please visit https://mail.cs.ualberta.ca/mailman/listinfo/MetaboNews
Back issues of this newsletter can be viewed from the newsletter archive (http://www.metabonews.ca/archive.html).
 |
1) Software Spotlight
|
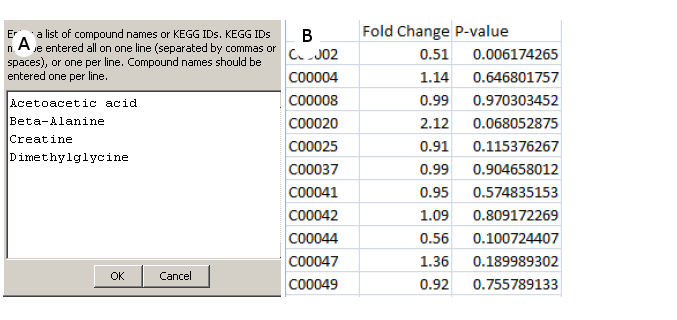
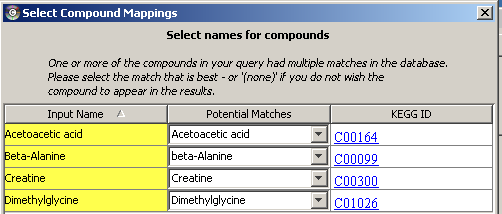
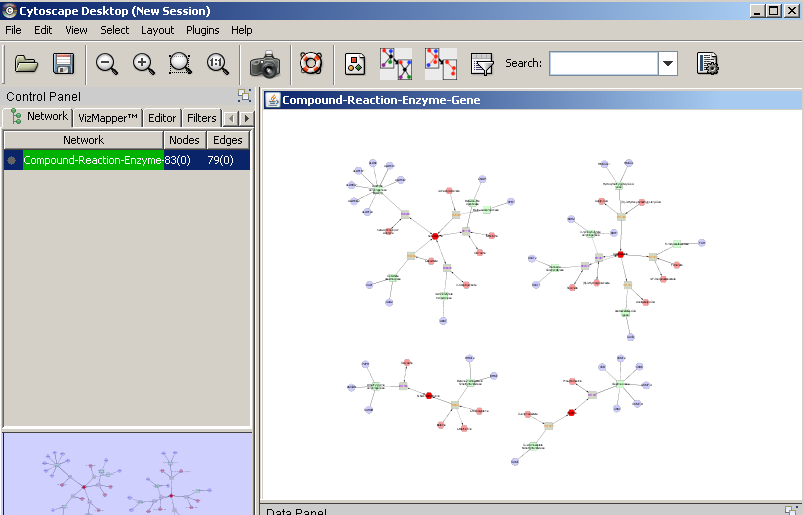
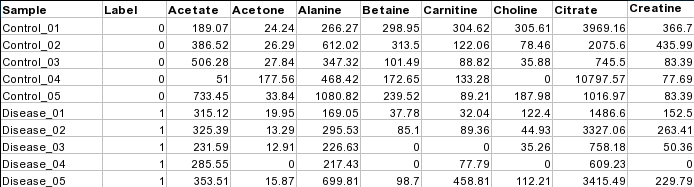
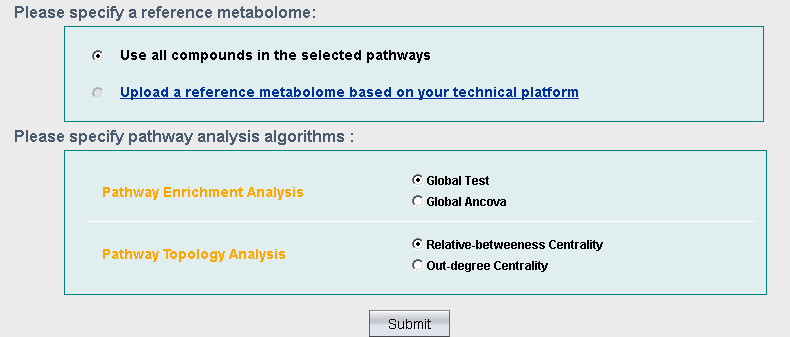
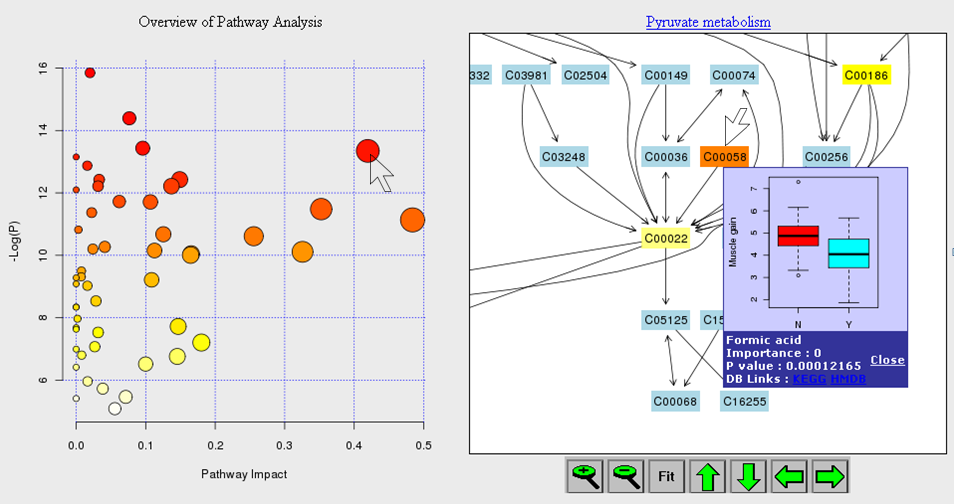
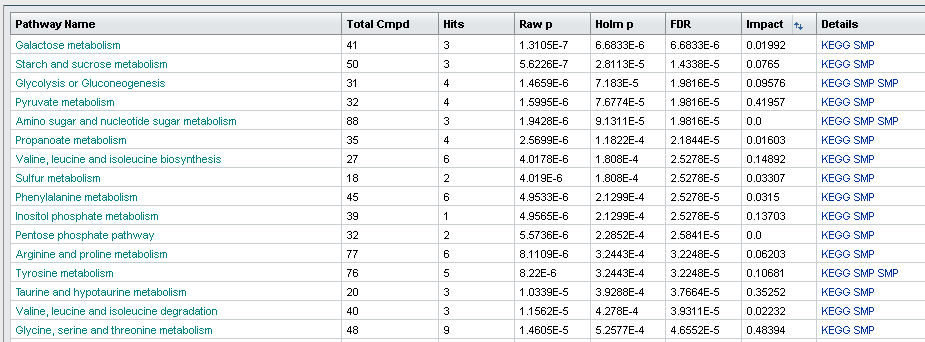
| |
2) Biomarker Beacon
|
 |
3) Metabolomics Current Contents
|
 |
4) MetaboNews |
| 26 Oct 2011 |
Study identifies genetic basis of human metabolic individuality - Led by researchers from Weill Cornell Medical College in Qatar and published in a recent edition of the journal Nature, the study provides details on the genetics behind many diseases, including cardiovascular and kidney disorders, diabetes, cancer, gout, thrombosis and Crohn's disease, and elucidates the role that individual differences in metabolism play in these disorders. Disturbances in metabolism are at the root of a variety of human afflictions and complex diseases. Although many of the genes that contribute to these conditions have been identified since the completion of the Human Genome Project in 2003, it is still not known how metabolic disorders related to these genetic aberrations disrupt cellular processes. Nature paper: Suhre K et al., Human metabolic individuality in biomedical and pharmaceutical research. Nature. 2011 Aug 31;477(7362):54-60. doi: 10.1038/nature10354. [PMID: 21886157] Source: Medical
Xpress
|
| 17 Oct 2011 |
Metabolon and Shanghai Jiao Tong University Open Metabolomics Lab in China - Metabolon, Inc., a diagnostics and services company offering the industry's leading biochemical profiling technology, announced today that it has opened a new metabolomics lab in Shanghai, China in collaboration with Shanghai Jiao Tong University (SJTU). The SJTU-Metabolon Joint Metabolomics Laboratory is a partnership with Shanghai Jiao Tong University, one of the oldest and most prestigious public research universities in China. Metabolon has licensed its proprietary metabolomics platform technology to SJTU to enable a world class biochemical profiling laboratory at the university. SJTU has funded the laboratory and personnel already trained by Metabolon and will work with academic and commercial scientists within China or other Asian countries to perform biochemical profiling experiments with Metabolon's technology. "We are very pleased to be bringing the science of metabolomics to a large untapped market such as China and the Asian continent as a whole," said Metabolon president and CEO John Ryals, Ph.D. "Our partnership with Shanghai Jiao Tong University is further proof of Metabolon's efforts to bring personalized medicine and metabolic biomarker research to all parts of the globe." Source: MarketWatch |
| 16 Oct 2011 |
Gut Bacteria May Keep Statin Response in Check - Various bacterial-derived bile acids appear to influence the response to statin treatment, researchers found. Pretreatment levels of several primary and secondary bile acids were strongly associated with the low-density lipoprotein (LDL) cholesterol-lowering ability of simvastatin in healthy individuals, according to Rima Kaddurah-Daouk, PhD, of Duke University, and colleagues. The findings, reported online in PLoS ONE, "warrant further evaluation of interactions of specific markers for gut microbiota and therapeutic response to statins," the researchers wrote. "Identification of the basis for such interactions may in turn lead to dietary or other interventions that can improve statin efficacy by altering gut microflora." There is variability among individuals in the therapeutic response to statins, and previous studies have shown that genetics can only account for part of it. Kaddurah-Daouk and colleagues turned to the field of metabolomics, which incorporates the interactions between a person's genome, microbiome, and environment in explaining response to treatments. PLoS ONE paper: Kaddurah-Daouk R et al., Enteric microbiome metabolites correlate with response to simvastatin treatment. PLoS One. 2011;6(10):e25482. Epub 2011 Oct 13. [PMID: 22022402] Source: MedPage Today |
 |
5)
Metabolomics Events
|
| 8-9 Nov 2011 |
7th annual Advances in Metabolic Profiling The conference will be co-located with Mass Spec Europe. Registered delegates will have access to both meetings ensuring a very cost-effective trip. For more information,
please visit http://www.selectbiosciences.com/conferences/AMP2011/.
|
| 13-16 Nov 2011 |
International Conference on Natural Products
(ICNP2011) - Metabolomics
A
New Frontier in Natural Products Science The conference is being organised by the Universiti Putra Malaysia and Malaysian Natural Products Society. The conference will be held at Palm Garden Hotel, IOI Resort, PUTRAJAYA from 13th -16th November 2011. For more information,
please visit http://www.icnp2011.upm.edu.my/.
|
| 20-22 Feb 2012 |
International Conference and Exhibition on
Metabolomics & Systems Biology Metabolomics-2012 will serve as a catalyst for the advances in the study of Metabolomics & Systems Biology by connecting scientists within and across disciplines at sessions and exhibition held at the venue, creates an environment conducive to information exchange, generation of new ideas, and acceleration of applications that benefit Research in Metabolomics & Systems Biology. Conference Highlights the following topics:
For more information,
please visit http://omicsonline.org/metabolomics2012/.
|
| 17-20 Apr 2012 |
analytica Conference 2012 In various symposiums, leading scientists from all over the world report on the latest developments, current trends and visions of the future. Analytic, diagnostic, biochemical and molecular biological methods and procedures are discussed here. On the last occasion, 140 well-known experts gave talks in 23 different thematic symposiums. Main subject emphases/highlights of the analytica Conference 2010
For more information,
please visit http://www.analytica.de/en/Home/congress/conference-program.
|
| 25-28 June 2012 |
METABOLOMICS 2012: Breakthroughs in plant,
microbial and human biology, clinical and
nutritional research, and biomarker discovery Local Organisers: Dan Bearden, Rick Beger, Rima Kaddurah-Daouk, Lloyd W. Sumner, Don Robertson, Padma Maruvada and Donna Kimball. For more information,
please visit http://www.metabolomics2012.org.
|
Please note: If you know of any metabolomics lectures, meetings, workshops, or training sessions that we should feature in future issues of this newsletter, please email Ian Forsythe (metabolomics.innovation@gmail.com).
 |
6) Metabolomics Jobs |
This is a resource for advertising positions in metabolomics.
If you have a job you would like posted in this newsletter,
please email Ian Forsythe (metabolomics.innovation@gmail.com).
Job postings will be carried for a maximum of 4
issues (8 weeks) unless the position is filled prior to that
date.
Jobs
Offered
| Job Title | Employer | Location | Date Posted | Source |
|---|---|---|---|---|
| Research
Associate |
Department of Biochemistry, University of Cambridge, UK |
Cambridge, UK |
24-Oct-2011 |
Metabolomics Society Jobs |
| Research
Scientist (Ph.D.) – Molecular Physiology - Metabolomics |
Algenol Biofuels | Bonita Springs, Florida |
13-Oct-2011 |
Metabolomics Society Jobs |
| Sr. Scientist /
Principal Scientist (R4-R5) – Analytical Chemistry – NMR |
Pfizer |
Groton, CT (New London/Norwich, Connecticut Area) |
10-Oct-2011 |
LinkedIn Jobs |
| Applications
Development Senior |
Seattle Children's Research Institute |
Greater Seattle Area | 6-Oct-2011 |
LinkedIn Jobs |
| Computational
Biologist - Metabolomics |
Genentech | South San Francisco,CA (San Francisco Bay Area) |
4-Oct-2011 |
LinkedIn Jobs |
| Business
Development Manager |
BASF – The Chemical Company |
Florham Park, New Jersey |
27-Sep-2011 |
Metabolomics Society Jobs |
| Assoc
Director - Bioinformatics |
Genentech |
United States |
16-Sep-2011 |
LinkedIn Jobs |
| Senior
Scientist 208819 (NCI) Job |
SAIC |
Frederick, MD, US (Washington D.C. Metro Area) |
12-Sep-2011 |
LinkedIn Jobs |
|
Ian J.
Forsythe, M.Sc.
MetaboNewsEditor Department of Computing Science
University of Alberta 221 Athabasca Hall Edmonton, AB, T6G 2E8, Canada Email: metabolomics.innovation@gmail.com Website: http://www.metabonews.ca |
This newsletter is produced by The
Metabolomics Innovation Centre (TMIC, http://www.metabolomicscentre.ca/) for
the benefit of the worldwide metabolomics community.
|