Metabolomics is a relatively new omics technology.
Both its analytical platforms and bioinformatics tools are
being rapidly developed. In this commentary, the author
briefly summarizes current concepts and progress in
metabolomic data analysis. The content is based mainly on the
lectures given by Dr. David Wishart during the Canadian
Bioinformatics Workshop on Informatics and Statistics for
Metabolomics (June 16-17, 2011, Edmonton, Canada).
Data normalization is necessary
There are two types of normalizations—biological
normalization and statistical normalization. Biological
normalization aims to reduce the biological variances unrelated
to the conditions of interest, e.g., variance introduced during
sample preparation (i.e., differences in sample volumes) or
different dilution factors in urine samples. Researchers
understand that it is necessary to address this difference
before proceeding to the next level of data analysis.
Statistical normalization aims to reduce the statistical
variances that may obscure the discovery of small, but
potentially significant signals. For instance, metabolite
concentrations usually span several orders of magnitude
(submicromolar to millimolar) and the effects from the highly
abundant metabolites tend to dominate. However, as we know, a
compound’s biological significance is not proportional to its
concentration. Therefore, it is important to perform appropriate
data transformations to make metabolites more comparable to each
other (i.e., to obtain a homogenous data set).
Data visualization is critical
Intuitive data visualization is critical for data
exploration and quality assurance. As metabolomic data usually
contains over hundreds of features, numerical summaries can
often hide important messages which can be easily picked up
through graphical presentation.
Fig.1
illustrates such a case. An outlier (indicated by a red arrow)
is obvious to any examiner. Is it caused by some measurement
error or typographical error? Addressing such an issue (i.e.,
through removal of the outlier) may significantly improve the
results of the downstream data analysis.
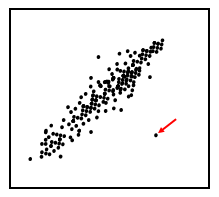
Figure 1. Data visualization for outlier detection.
Combining univariate and multivariate methods to achieve
better understanding
Identification of significant metabolites is usually the first
step towards finding useful biomarkers or identifying associated
biological processes. Univariate approaches are widely used in
this regard. Many well-established approaches (e.g., t-tests,
ANOVA) are available. The main advantages of univariate methods
are easy to understand and support for very flexible
experimental designs.
Multivariate statistics involves the simultaneous analysis of
more than two statistical variables. Because metabolomic data
usually contains hundreds of metabolite concentrations,
multivariate data analysis is considered ideal for such data
sets. The most widely used multivariate methods in metabolomics
are probably principal component analysis (PCA) and partial
least squares discriminant analysis (PLS-DA). Both of them are
capable of projecting high-dimensional data into two or three
dimensions that can be conveniently visualized for pattern
discovery. PCA is an unsupervised method, suitable to see if
natural clusters form or if data separates well.
Fig.
2 illustrates a PCA result which forms three natural
clusters. PLS-DA is a supervised method that uses group
information to help improve the class separation and is often
used when PCA shows little separation.
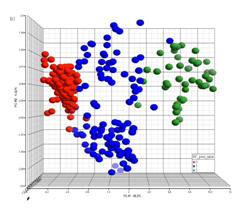
Figure 2. PCA visualization of natural clustering
pattern.
Is there an overall significant difference between the
metabolomic profiles?
For univariate analysis such as t-tests, researchers can
judge whether a given metabolite is significant based on its
p-value. For multivariate approaches, calculating a p-value is
difficult as we do not know the characteristics of the
underlying distribution. However, we can calculate an empirical
p-value using a non-parametric approach such as a permutation
test. This approach involves randomly reassigning the class
labels and performing the analysis on the newly labeled data
set. The process is repeated hundreds or thousands of times and
the performance measures are plotted on a histogram for visual
assessment. From the resulting histogram it is possible to
determine if the of the original class assignment is
significantly different from or a part of the distribution based
on the permuted class assignments. The empirical p-value is
calculated as the proportion of times that the permuted data
yielded a better result than the one using the original labels.
For example, if none of the permuted classes is better than the
observed one in 2000 permutations, the p-value is reported as
p<0.0005 (less than 1/2000). With more permutations, the
empirical p-value will be very close to the real p-value. The
process is illustrated in
Fig.
3.
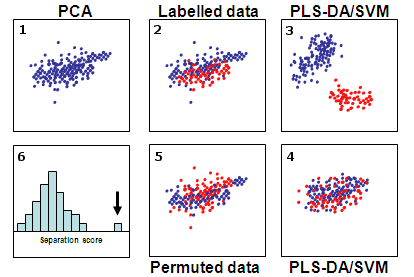
Figure 3. Permutation
tests to evaluate the significance of separation. Panel 1 shows
the overview by principal component analysis (PCA). Panel 2
shows the same PCA result colored by group labels. Only slight
separation can be seen between the two groups. Panel 3 shows the
supervised methods PLS-DA/SVM (partial least
squares-discriminant analysis/support vector machines) on the
same dataset with much better separation. Panels 4 and 5 shows
the separations using the permuted datasets. The histogram in
Panel 6 shows the comparison between the results using permuted
datasets and the one using original data (indicated by a black
arrow). In this case, the separation is clearly significant.
Conclusions
Metabolomics is a recent addition to the omics family.
There are certain advantages to being the last to arrive. Many
of the statistical and computational methods are borrowed from
other omics platforms and adapted to the specific needs of
metabolomic data, allowing metabolomics to quickly catch up to
its more mature cousins. Another important source of concepts
grew from the field of chemometrics such as the use of PCA and
PLS-DA. Most of the approaches discussed here are freely
available online through the MetaboAnalyst (
http://www.metaboanalyst.ca)
web application.









