Phenol-Explorer, a database on dietary polyphenols and their
metabolites
Feature article contributed by Augustin Scalbert, Head
of Biomarkers Group, International Agency on Research on
Cancer (IARC), Lyon, France
Foods are derived from
various animal and plant species and each species is characterized
by the presence of large number of chemical constituents. Some of
these constituents have received considerable attention due to
their established or suggested role in nutrition and health. These
include polyphenols abundant in wine, tea, coffee, fruits,
wholegrain cereals, and many other foods of plant origin. They
have become very popular because of their antioxidant properties
and their possible role in the prevention of diseases, such as
cardiovascular diseases, diabetes, cancers, osteoporosis, and
neurodegenerative diseases.
Polyphenols constitute one of the largest families of
phytochemicals with over 500 compounds described in various foods.
We developed Phenol-Explorer, the first comprehensive database on
dietary polyphenols, which describes 502 polyphenols in 452 foods
(1) (
http://www.phenol-explorer.eu/).
The database has been used to measure polyphenol intake in
populations and to study associations between intake and disease
risk (2).
Once ingested with our diet, polyphenols are rapidly absorbed and
metabolized, either by the gut microbiota or in our tissues. A
detailed knowledge of the bioavailability of the various
polyphenols and the nature and concentrations of the metabolites
formed in the body is also essential to understand their effects
on health. We recently released the second version of
Phenol-Explorer (Phenol-Explorer 2.0) which now includes 375
polyphenol metabolites described in the literature (
http://www.phenol-explorer.eu/metabolites)
together with detailed pharmacokinetic data (3) (
Figure
1). It contains descriptions of the animal or human studies
(food or polyphenol ingested, dose administered, composition of
the administered food, etc.) where the compounds were observed,
with links to the original literature source.
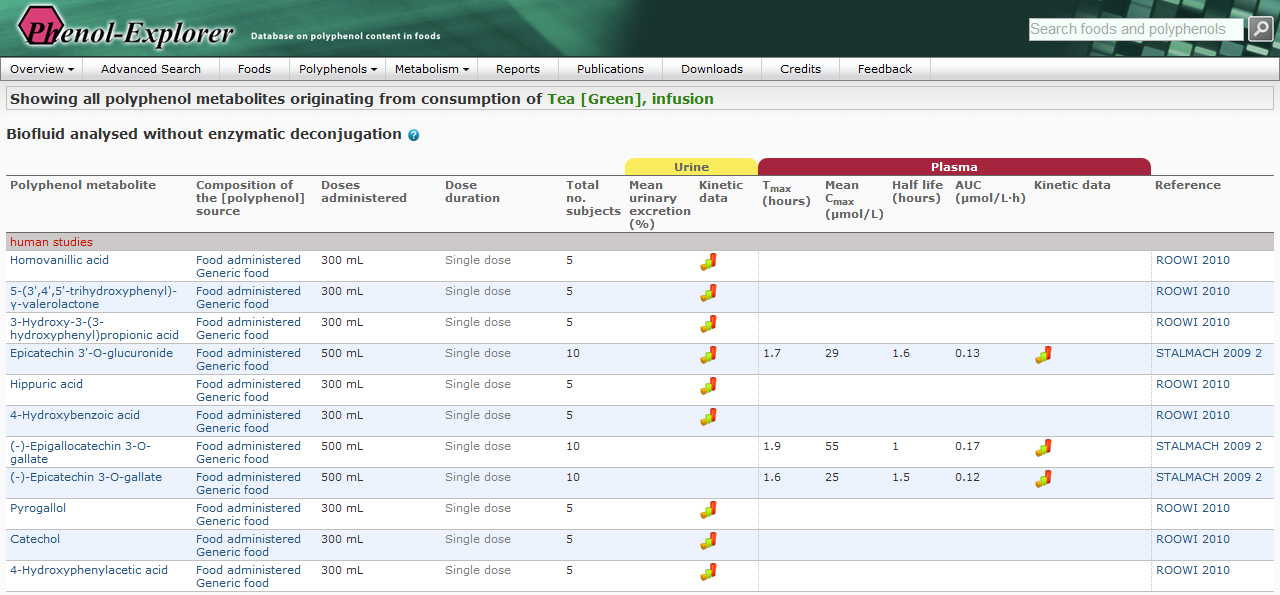
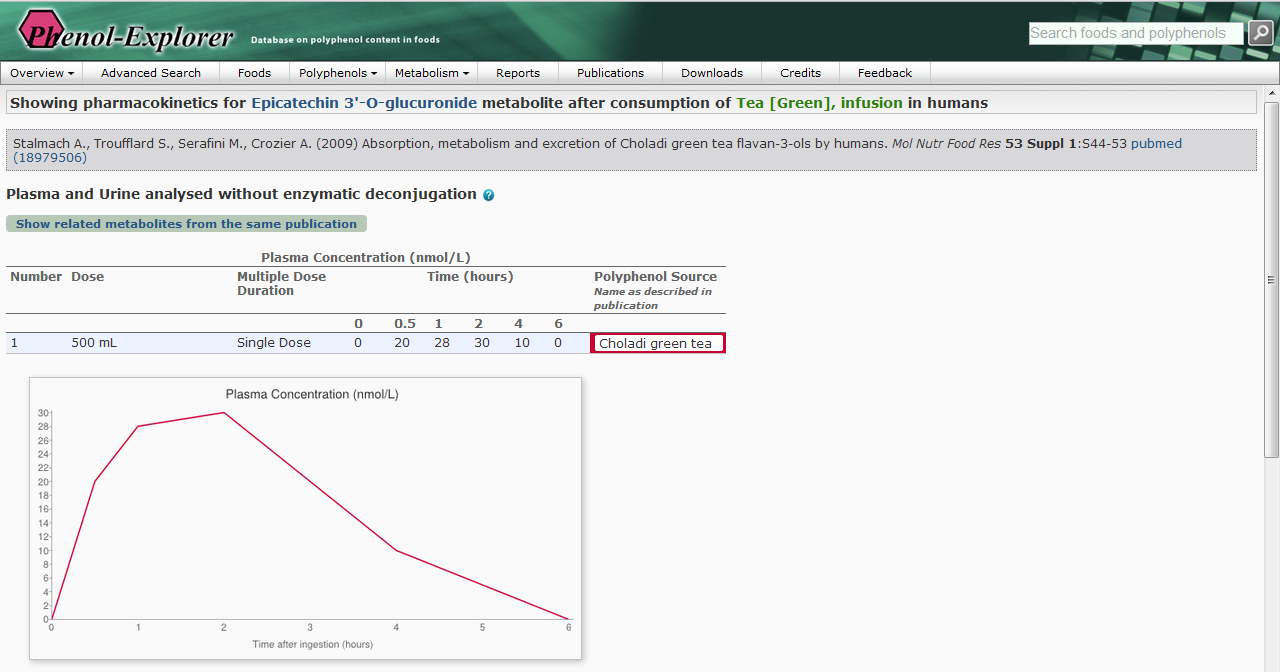 Figure 1.
Figure 1. Screenshots of the Phenol-Explorer database
showing metabolites formed after ingestion of green tea (above)
together with their pharmacokinetic properties (below).
Each metabolite is described with its molecular formula, chemical
structure, molecular weight, and links to other databases such as
CAS, ChEBI, and PubChem (
Figure
2).
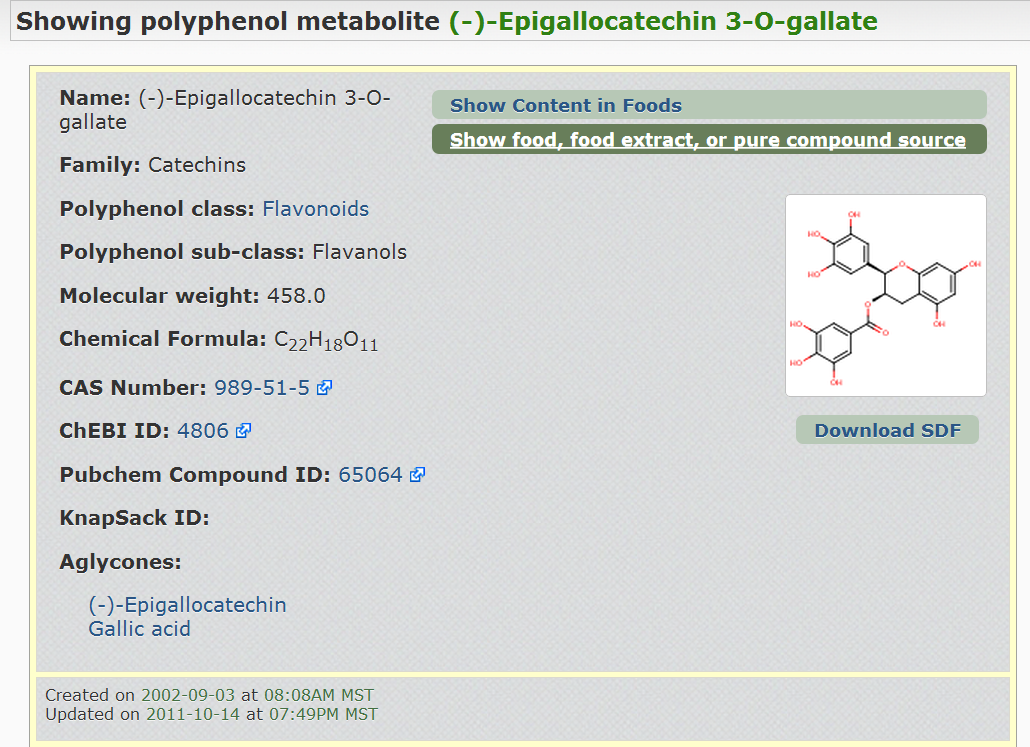 Figure 2.
Figure 2. Screenshot of the Phenol-Explorer database
showing the structure of a metabolite found after tea ingestion.
Various queries can be made in Phenol-Explorer 2.0 to identify all
metabolites formed from a particular polyphenol. For example, the
database allows users to retrieve the 27 metabolites formed from
chlorogenic acid, the main polyphenol present in coffee, in rat
and human feeding studies (
http://www.phenol-explorer.eu/metabolism/polyphenol-metabolites/948),
or polyphenol metabolites identified in urine or plasma after
consumption of coffee (
http://www.phenol-explorer.eu/metabolism/metabolites/1089).
All data and results of query searches can be exported as CSV or
Excel files.
This information on polyphenol metabolites is now used in our lab
to identify in complex urine fingerprints from free-living
subjects all polyphenol metabolites, which may serve as biomarkers
of polyphenol exposure for future epidemiological studies (
Figure
3).
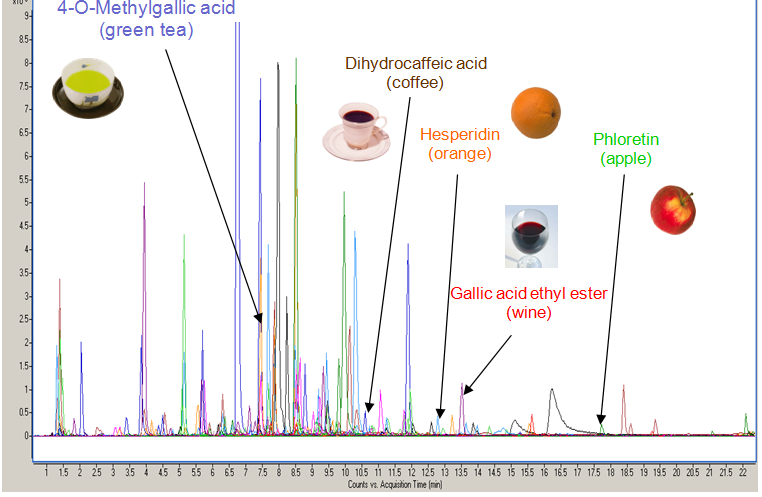 Figure 3.
Figure 3. Urinary profile obtained by high-resolution mass
spectrometry (UPLC-QTof-MS) showing 85 putative polyphenol
metabolites identified with the Phenol-Explorer database.
Phenol-Explorer is the first database describing in a systematic
way all metabolites formed from a particular class of food
compounds. This database can be particularly useful to identify
metabolites formed from dietary polyphenols in human biofluids
which constitute altogether a significant fraction of what we have
called the 'food metabolome' (4).
Acknowledgments
Phenol-Explorer 2.0 has been developed at INRA, Clermont-Ferrand
(J. Rothwell, C. Manach, A. Scalbert) and IARC, Lyon (Will
Edmands, A. Scalbert) in collaboration with the University of
Barcelona (M. Urpi-Sarda, M. Boto-Ordonez, R. Llorach, C.
Andres-Lacueva), the University of Alberta, Edmonton (D. Wishart,
V. Neveu), and Insiliflo, Edmonton (C. Knox), with the financial
support of Danone Research and the Institut National du Cancer,
Paris, France.
References
- Neveu V, Perez-Jiménez J, Vos F, Crespy V, du
Chaffaut L, Mennen L, Knox C, Eisner R, Cruz J, Wishart D,
Scalbert A. Phenol-Explorer: an online comprehensive database
on polyphenol contents in foods. Database (Oxford).
2010;2010:bap024. Epub 2010 Jan 8. [PMID:
20428313]
- Pérez-Jiménez J, Fezeu L, Touvier M, Arnault
N, Manach C, Hercberg S, Galan P, Scalbert A. Dietary intake
of 337 polyphenols in French adults. Am J Clin Nutr. 2011
Jun;93(6):1220-8. Epub 2011 Apr 13. [PMID:
21490142]
- Rothwell JA, Urpi-Sarda M, Boto-Ordoñez M, Knox C,
Llorach R, Eisner R, Cruz J, Neveu V, Wishart D, Manach C,
Andres-Lacueva C, Scalbert A. Phenol-Explorer 2.0: a major
update of the Phenol-Explorer database integrating data on
polyphenol metabolism and pharmacokinetics in humans and
experimental animals. Database (Oxford). 2012 Aug
9;2012:bas031. [PMID:
22879444]
- Manach C, Hubert J, Llorach R, Scalbert A. The complex links
between dietary phytochemicals and human health deciphered by
metabolomics. Mol Nutr Food Res. 2009 Oct;53(10):1303-15.
Review. [PMID:
19764066]
Please
note: If you know of any
metabolomics research programs, software, databases,
statistical methods, meetings, workshops, or training
sessions that we should feature in future issues of this
newsletter, please email Ian Forsythe at metabolomics.innovation@gmail.com.