MUSCLE—Multi-Platform Unbiased Optimisation of Spectrometry
via Closed Loop Experimentation
Feature article contributed by Mark Viant1,
Shan He1, Warwick Dunn1, Gregory
Genta-Jouve1, James Bradbury1, Joshua
Knowles2, Roy Goodacre2
1. University of Birmingham, Birmingham, UK and 2. University of
Manchester, Manchester, UK
Liquid chromatography mass spectrometry (LC-MS) is an extremely
widely used tool in analytical laboratories for selectively
measuring a broad range of chemicals. It is also the primary
technology in metabolomics. However the development of new LC-MS
methods, or even the transfer of an existing method between
instruments and laboratories, is both time consuming and
challenging and places a considerable burden on the analyst. This
bottleneck significantly impedes our ability to establish new
bioanalytical methods, e.g., for the targeted analysis of a
particular metabolite class or metabolic pathway. The primary
reason why optimising an LC-MS method is so challenging stems from
the large number of instrument control settings. Varying all of
the LC and MS parameters systematically to optimise an analysis is
simply impossible because of the astronomical number of
combinations that are possible.
Previously we developed a closed-loop strategy for the automated
optimisation of metabolite analyses on three platforms: LECO
GC-ToF-MS and GCxGC-ToF-MS, and Waters LC-ToF-MS [
1-3].
Not only was this procedure fully automated, but it greatly
improved the analytical method by detecting three times as many
metabolites, and it only took a few days of automated optimisation
to achieve this result. However, since it was programmed to
control only three specific mass spectrometers, scientists have
not been able to deploy this automated optimisation software in
their own research laboratories. Also, because of how we wrote our
original software, reprogramming it for each additional mass
spectrometer would be challenging and time consuming.
With funding from the UK Biotechnology and Biological Sciences
Research Council, we have developed and validated a
multi-platform, user-friendly software tool, MUSCLE, for the
robust, objective and automated optimisation of targeted LC-MS/MS
analyses (
Figure
1) [
4].
Click on the thumbnail below to view a larger version of the
image
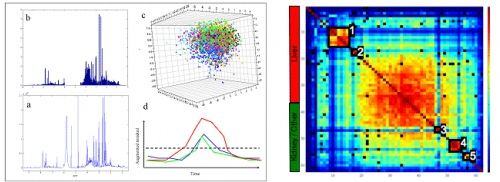
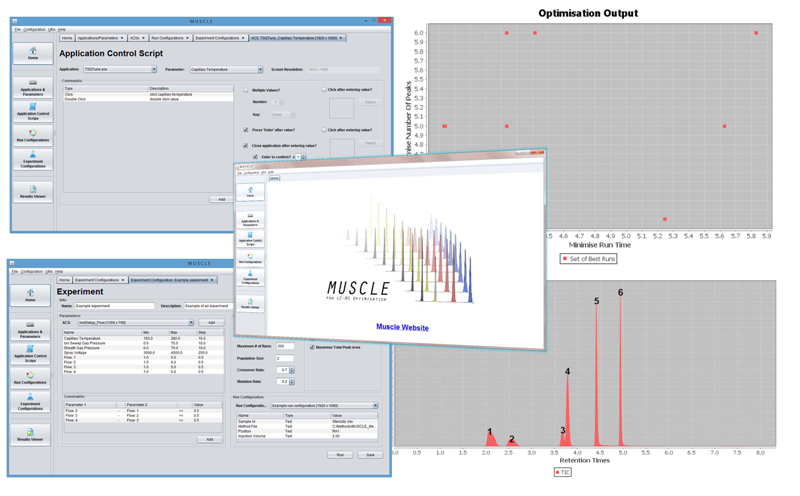 Figure 1. MUSCLE screenshots: Top Left:
Figure 1. MUSCLE screenshots: Top Left: Experiment
setup,
Bottom Left: Application Control Script (ACS)
setup,
Top Right: Output of an optimisation,
Bottom
Right: Chromatogram with peak detection,
Middle: Homepage.
MUSCLE can be used to optimise methods on a wide array of LC-MS/MS
platforms in a fully automated manner. Our approach is based on
utilising the power of genetic algorithms and closed-loop
optimisation to discover optimal solutions of LC and MS
parameters, readily enabling analysts with no programming skills
to optimise their targeted LC-MS/MS analyses. With user-defined LC
and MS parameter selections, as well as user-defined criteria for
spectral quality, MUSCLE can increase the speed and quality of
one's method development regardless of the instrument
manufacturer. Application of this software on a Thermo Scientific
TSQ Vantage and Waters Xevo™ TQ, for the analysis of a steroid
mixture, decreased the run time and increased the sensitivity of
analysis as indicated below (
Figure
2).
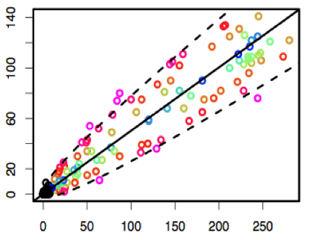
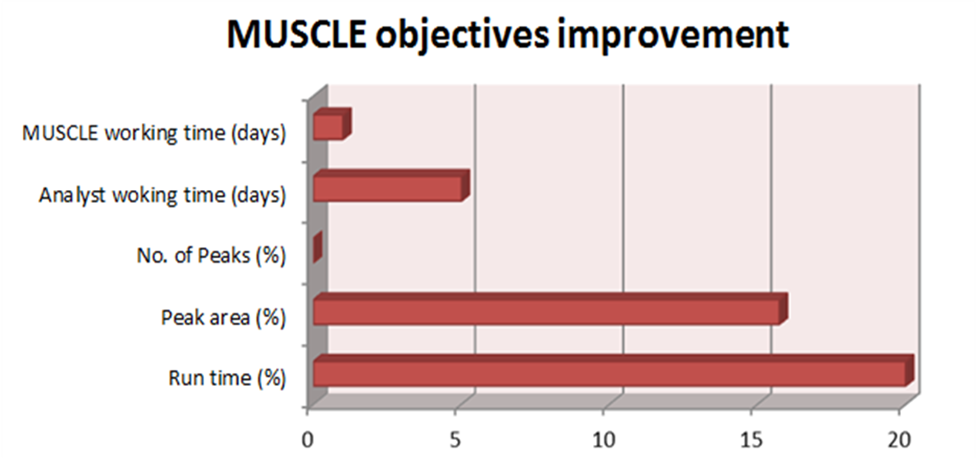 Figure 2. Optimisation results:
Figure 2. Optimisation results: Following an optimisation
study that comprised of 170 LC-MS/MS runs, MUSCLE had reduced the
run time of the LC method by 20%, while simultaneously increasing
the total peak area by 15%. This was all achieved in 28 hours of
fully automated optimisation. We estimate that to achieve
comparable method optimisation manually would take an analyst 5
full days of manual work.
Could this be of benefit to you?
There are direct and obvious benefits of MUSCLE to scientists who
employ LC-MS/MS. Novel analytical method development, the
re-implementation of published methods to a new laboratory, and
the transfer of methods across instruments within the same
laboratory will become easier, more rapid, and less expensive. In
turn this could improve the quality of analysis, and will
facilitate the analyst to attempt method development and
optimisation that they could not previously consider.
Where can I get MUSCLE?
The beta release of MUSCLE will be available in September 2013 for
download at
http://www.muscleproject.org/,
where you can also find training documentation and videos. We are
now seeking testers across academia, industry, and government
laboratories.
If you are interested in beta testing or have any inquiries,
please contact
info@muscleproject.org.
To be kept up to date with all the latest MUSCLE news, please
register your interest directly at
http://www.muscleproject.org/.
The developers
MUSCLE was developed by a team of computer scientists, analytical
chemists, and biochemists from the Universities of Birmingham and
Manchester in the UK, including Professor Mark Viant and Drs. Shan
He, Warwick Dunn, Gregory Genta-Jouve and Mr. James Bradbury from
Birmingham, and Dr. Joshua Knowles and Professor Roy Goodacre from
Manchester.
References
[1] S O'Hagan, WB Dunn, M Brown, JD Knowles, DB Kell,
Closed-loop, multiobjective optimization of analytical
instrumentation: gas chromatography/time-of-flight mass
spectrometry of the metabolomes of human serum and of yeast
fermentations. Anal. Chem. 77: 290-303 (2005). [PMID:
15623308]
[2] S O'Hagan, WB Dunn, JD Knowles, D Broadhurst, R Williams, JJ
Ashworth, M Cameron, DB Kell, Closed-loop, multiobjective
optimization of two-dimensional gas chromatography/mass
spectrometry for serum metabolomics. Anal. Chem. 79: 464-476
(2007). [PMID:
17222009]
[3] E Zelena, WB Dunn, D Broadhurst, S Francis-McIntyre, KM
Carroll, P Begley, S O'Hagan, JD Knowles, A Halsall, ID Wilson,
DB Kell, Development of a robust and repeatable UPLC-MS method
for the long-term metabolomic study of human serum. Anal. Chem.
81: 1357-1364 (2009). [PMID:
19170513]
[4] J Bradbury, G Genta-Jouve, WB Dunn, S O’Hagan, R Goodacre,
JD Knowles, MR Viant, MUSCLE: A novel multi-platform,
user-friendly software tool for the robust, objective and
automated optimisation of targeted LC-MS analyses. 9th Annual
International Conference of the Metabolomics Society, 1-4 July
2013, Glasgow, UK.
Please
note: If you know of any
metabolomics research programs, software, databases,
statistical methods, meetings, workshops, or training
sessions that we should feature in future issues of this
newsletter, please email Ian Forsythe at metabolomics.innovation@gmail.com.