Customized Metabolomics Databases for the Analysis
of TOCSY-Type NMR of Complex Mixtures
Feature article contributed by Kerem Bingol and Rafael Brüschweiler
Department of Chemistry and Biochemistry, The Ohio State
University, Columbus, Ohio 43210, United States
Campus Chemical Instrument Center, The Ohio State University,
Columbus, Ohio 43210, United States
Compound identification of complex mixtures by 2D TOCSY-based
NMR spectroscopy using common 1D NMR spectral databases is a
challenge because magnetization transfers in TOCSY experiments are
limited to spins within the same spin system. This situation
frequently leads to unexplained peaks in the 1D NMR spectrum of a
metabolite if they belong to different spin systems. To address
this, we recently developed NMR metabolomics spectral databases,
which were specifically designed to deal with this type of
situation.
Our customized metabolomics NMR databases, termed
1H(
13C)-TOCCATA [
1]
and
13C-TOCCATA [
2],
contain complete
1H and
13C chemical shift information of
individual spin systems and isomeric states of over 450 common
metabolites. Since this information is directly reflected in
cross-sections of 2D
1H-
1H TOCSY, 2D
13C-
1H HSQC-TOCSY, and 2D
13C-
13C
TOCSY spectra, these databases allow the accurate and
straightforward identification of metabolites of complex metabolic
mixtures from these types of experiments.
The
1H(
13C)-TOCCATA
database is best suited for 2D
1H-
1H TOCSY and 2D
13C-
1H HSQC-TOCSY spectra of complex
mixtures at
13C natural
abundance [
1],
whereas the
13C-TOCCATA is best
applied to 2D
13C-
13C constant-time (CT) TOCSY spectra
of uniformly
13C labeled
samples [
2].
Both databases can be accessed through public web portals:
1H(
13C)-TOCCATA
at
http://spin.ccic.ohio-state.edu/index.php/toccata2/index
and
13C-TOCCATA at
http://spin.ccic.ohio-state.edu/index.php/toccata/index.
We illustrate the performance of
1H(
13C)-TOCCATA for the 2D
1H-
1H
TOCSY spectrum of a cell lysate from
E. coli (
Figure
1), which yields a substantial improvement over other
databases as well as 1D
1H
NMR-based approaches in the number of compounds that can be
correctly identified with high confidence. This database can be
equally well applied other biological samples, such as urine,
blood, and tissue extracts.
Click on thumbnail below to view a larger version of the image
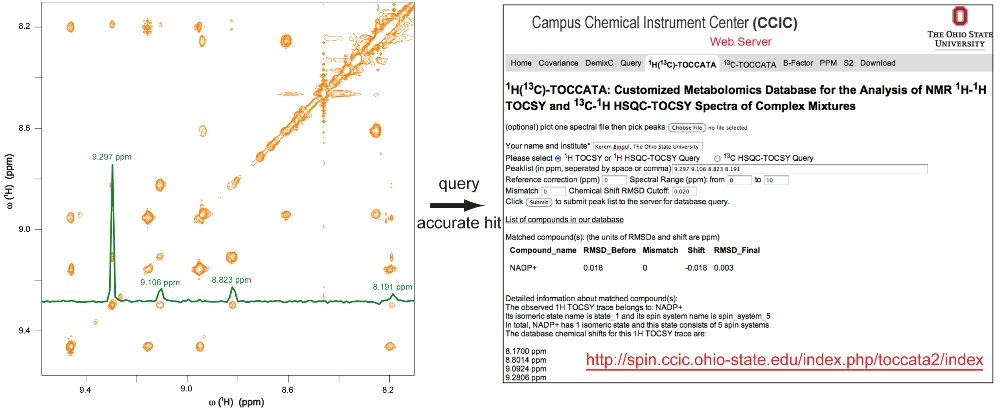 Figure 1.
Figure 1. Schematic representation of the querying
approach using the new
1H(
13C)-TOCCATA database. In the 2D
1H-
1H
TOCSY spectrum of an
E. coli cell lysate (orange), the
1H TOCSY trace (green cross-section)
is extracted and its peaks are queried against the new database.
The
1H TOCSY trace is assigned
with high accuracy to the only viable hit, which is the
nicotinamide ring portion of NADP
+.
The green
1H TOCSY trace of the
orange spectrum in
Figure
1 was directly searched against
1H(
13C)-TOCCATA. The search returned a
single hit, which is the correct one corresponding to the
nicotinamide ring of NADP
+. NADP
+ is a large metabolite with several
substructures, such as adenine, ribose rings, and nicotinamide.
Since in a conventional 1D
1H
NMR database, the peaks of all parts of the molecule are
represented as a single group, searching the peaks of only the
nicotinamide ring against the 1D
1H
NMR database will create mismatches, which precludes accurate
identification. By contrast, with the help of the new
1H(
13C)-TOCCATA
database, one can easily identify individual spin systems
(substructures) and isomeric states of metabolites with optimal
accuracy.
Citing TOCCATA
[1]
1H(
13C)-TOCCATA
is a free and publicly accessible web portal. The link to the
database is:
http://spin.ccic.ohio-state.edu/index.php/toccata2/index
Publication:
Kerem Bingol, Lei Bruschweiler-Li, Da-Wei Li and Rafael
Brüschweiler. Customized Metabolomics Database for the Analysis of
NMR
1H-
1H
TOCSY and
13C-
1H
HSQC-TOCSY Spectra of Complex Mixtures. Analytical Chemistry, 86,
5494-5501 (2014).
http://pubs.acs.org/doi/abs/10.1021/ac500979g
[2] 13C-TOCCATA is a free
and publicly accessible web portal. The link to the database is:
http://spin.ccic.ohio-state.edu/index.php/toccata/index
Publication:
Kerem Bingol, Fengli Zhang, Lei Bruschweiler-Li, and Rafael
Brüschweiler. TOCCATA: A Customized Carbon Total Correlation
Spectroscopy NMR Metabolomics Database. Analytical Chemistry,
84, 9395–9401 (2012). http://pubs.acs.org/doi/abs/10.1021/ac302197e
Please
note: If you know of any
metabolomics research programs, software, databases,
statistical methods, meetings, workshops, or training
sessions that we should feature in future issues of this
newsletter, please email Ian Forsythe at metabolomics.innovation@gmail.com.