
TMIC and the Metabolomics Society
Issue 72 - August 2017
CONTENTS:
Online version of this newsletter:
http://www.metabonews.ca/Aug2017/MetaboNews_Aug2017.htm


 |
| Published
in partnership between TMIC and the Metabolomics Society Issue 72 - August 2017 |
|
CONTENTS: |
|
|
 |
| TMIC
Services |
 |
| npj
Molecular Phenomics: Call for Papers |
 |
Metabolomics Society News |
and
| |
Metabolomics
Spotlight
|
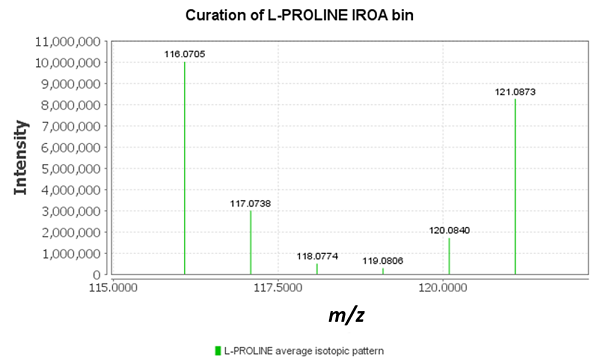
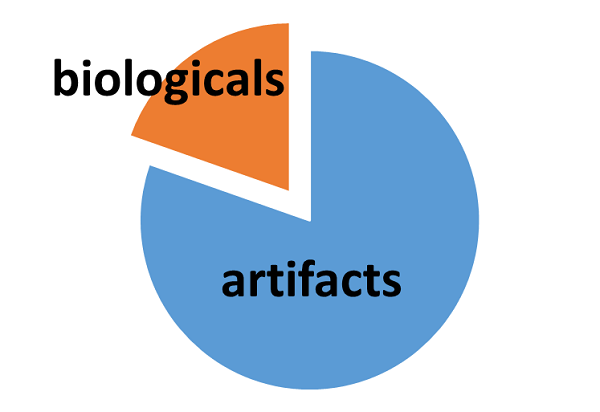
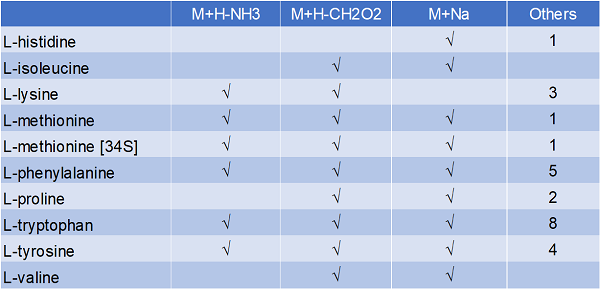
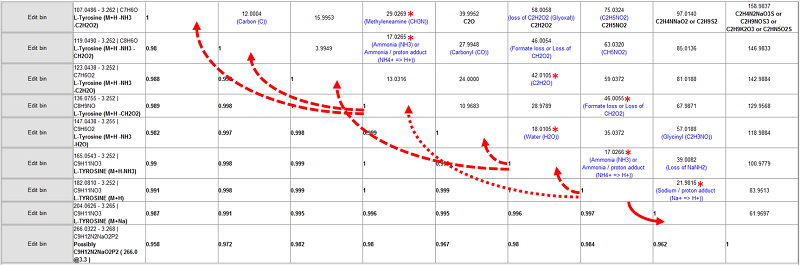
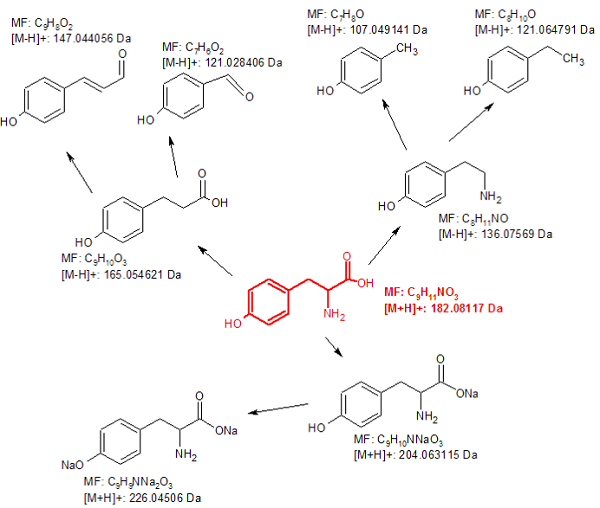
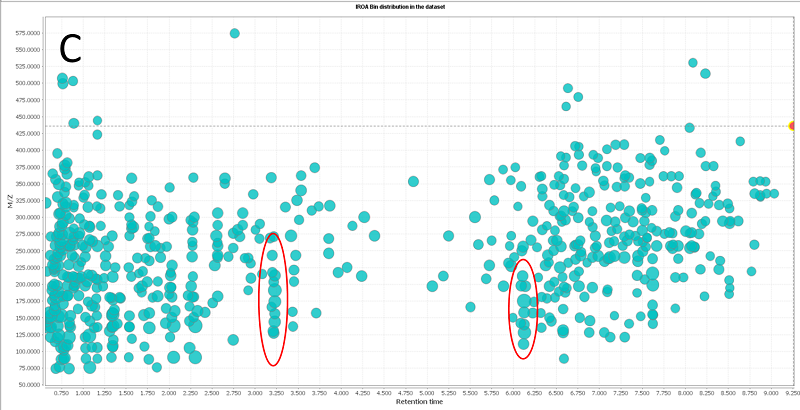
The IROA-Matrix has been developed into a Workflow Kit which includes Standards and software. Further information can be freely found here and the Kit User Manual obtained from the primary author of this article.
References
| |
MetaboInterview
|
| PhD
Student, Wishart Research Group, University of
Alberta, Edmonton, Canada |
|
 |
Biography Seyed Ali obtained his BSc degree in Animal Science from the College of Agriculture & Natural Resources, University of Tehran, I.R. Iran, and his MSc in Nutritional Immunology from the University of Alberta. He is currently conducting research in an interdisciplinary field looking into the identification of predictive biomarkers of sheep feed efficiency, carcass merit, pregnancy and litter size using different "omics" technologies, with a main focus on metabolomics. |
 |
Metabolomics
Current Contents
|
Metabolomics
Events
|
| 23-24
Aug 2017 |
Swedish Metabolomics Conference 2017: Metabolomics and Health Venue: Chalmers University of Technology, Gothenburg, Sweden Confirmed speakers:
|
| 11-15
Sep 2017 |
School on "Cloud-based Metabolomics Data Analysis and Collaboration" Venue: Building 2 of the Technology Park of Sardinia Pula, Sardinia, Italy We are happy to announce that registrations are now open; visit http://cloudmet2017.crs4.it/ to register now. Metabolomics is a well established -omics science whose growth is bringing about new challenges. Systematic studies and integration with other data sources are resulting in ever larger dataset sizes; production applications require superior computational scalability of analysis techniques; complex, multi-step workflows make study reproducibility more challenging. At the same time, cloud computing technologies are extending their functionality and provide practical solutions for many of these problems. In this School students will have the opportunity to learn about current topics in metabolomics, with a slant on the integration of cloud computing technologies where they are beneficial to the effectiveness and efficiency of research and analysis work. Top-level lecturers in the field will provide their insight and will be available for the entire duration of the school, with ample opportunity for interaction with the students. Importantly, the School will also include practical sessions where students can put their new knowledge into practice under the guidance of tutors and run analyses using the new PhenoMeNal cloud-based metabolomics platform. To register and for more information, visit http://cloudmet2017.crs4.it/. |
| 20-23 Sep
2017 |
MOVISS: Bio and Data Venue: Vorau, Austria A problem driven meeting aimed at bioinformaticians, biochemists, statisticians and those who handle and interpret metabolomics data When: Sept 20-23, 2017 Where: Vorau, Austria To register and for more information, go to www.MOVISS.eu, and follow us on Twitter @MOVISSmeet Not just an ordinary conference, where people present work they have already done, MOVISS is centered on identifying and problem solving current challenges relating to metabolomics data handling by getting everyone in the room discussing it. Want to be at the forefront of solving some of the major bottlenecks in the Metabolomics Revolution – see you at MOVISS! A follow up R Summer School will be held on September 25-27, 2017 in Vorau, Austria. |
| 25-27
Sep 2017 |
Joint OpenMS and MetFrag User Meeting Venue: Martin-Luther University, Halle, Germany Speakers include:
|
| 2
Oct 2017 |
Metabolomics: Understanding Metabolism in the 21st Century FREE online course Metabolomics is an emerging field that aims to measure the complement of metabolites (the intermediates and products of metabolism) in living organisms. In this course, we provide an introduction to metabolomics and describe the current challenges in analysing the complement of metabolites in a biological system. The course is targeted towards final year undergraduate students from biology / chemical disciplines and medical students, but will also provide a valuable introduction to the metabolomics field for MSc and PhD students, and scientists at any stage in their careers. Register at: https://www.futurelearn.com/courses/metabolomics or email: bmtc@contacts.bham.ac.uk for more information. |
| 12-13
Oct 2017 |
Quality Assurance and Quality Control in Metabolomics Venue: Birmingham Metabolomics Training Centre, School of Biosciences, University of Birmingham, Birmingham, UK This two-day course will provide a comprehensive overview of the application of quality assurance (QA) and quality control (QC) in metabolic phenotyping. The course is aimed at students and researchers who are actively working in the field. Experts who have developed the application of QA and QC procedures within the field will lead the course. It will include both theoretical and practical components to:
Bursaries are now available for PhD students funded by the BBSRC, MRC, or NERC. For further information and registration details, please visit http://www.birmingham.ac.uk/facilities/metabolomics-training-centre/courses/quality-phenotyping.aspx or contact bmtc@contacts.bham.ac.uk. |
| 12-13
Oct 2017 |
Mayo Clinic Metabolomics Symposium Venue: Leighton Auditorium, Siebens Building, Mayo Clinic, Rochester, Minnesota, USA Mayo Clinic will host a Metabolomics Symposium from October 12 to October 13 on the Rochester, Minn., campus. The event will feature presentations on the practice and theory of metabolomics applications, the latest research in metabolomics and networking opportunities. The workshop is open to beginning and established researchers, students and postdoctoral fellows. A focused workshop is available to a limited group of individuals based on availability. Scholarships for scholars and junior faculty are available. For more information, please visit http://matrix.conferenceataclick.com/metabolomics_2017/home_live1.htm. |
| 18-20
Oct 2017 |
Workshop in Environmental Omics, Integration and Modelling Venue: CosmoCaixa, Barcelona, Spain The aim of this workshop is to review the state-of-the-art and broaden the knowledge on high-throughput analytical methods, data integration and modelling in Environmental Omics and Toxicology. The main workshop topics will be the following:
For more information, please visit http://wenvomics2017.info/. |
| 6-8
Nov 2017 |
Metabolomics with the Q Exactive Venue: Birmingham Metabolomics Training Centre, School of Biosciences, University of Birmingham, Birmingham, UK This three-day course will introduce you to using the Q Exactive mass spectrometer in your metabolomics investigations. The course is aimed at students and researchers with minimal previous experience of applying LC-MS in metabolomics. The course will be led by experts in the field and include lectures, laboratory sessions and computer workshops to provide:
Bursaries are now available for PhD students funded by the BBSRC, MRC, or NERC. For further information and registration details, please visit http://www.birmingham.ac.uk/facilities/metabolomics-training-centre/courses/q-exactive.aspx or contact bmtc@contacts.bham.ac.uk. |
| 1
Dec 2017 |
Introduction to Metabolomics for the Clinical Scientist Venue: Birmingham Metabolomics Training Centre, School of Biosciences, University of Birmingham, Birmingham, UK This one-day course in partnership with the Phenome Centre Birmingham will provide clinicians with an overview of the metabolomics pipeline, highlighting the benefits of the technique to the medical field. The course will provide an:
For further information and registrations details, please visit http://www.birmingham.ac.uk/facilities/metabolomics-training-centre/courses/introduction-metabolomics.aspx or contact bmtc@contacts.bham.ac.uk. |
| 6-8
Dec 2017 |
Multiple biofluid and tissue types, from sample preparation to analysis strategies for metabolomics Venue: Birmingham Metabolomics Training Centre, School of Biosciences, University of Birmingham, Birmingham, UK This 3-day course will provide a comprehensive overview of dealing with complex biological samples for LC-MS analysis. The course is targeted towards students and researchers who are actively applying metabolomics in their research. The course will be led by experts in the field and include:
For further information and registration details, please visit http://www.birmingham.ac.uk/facilities/metabolomics-training-centre/courses/sample-analysis.aspx or contact bmtc@contacts.bham.ac.uk. |
| 11-13
Dec 2017 |
MetaboMeeting 2017 Venue: University of Birmingham, UK Make plans to attend the 10th successful MetaboMeeting conference. The meeting will bring together research scientists and practitioners from all areas of application and development of metabolic profiling, covering a wide range of experience from early career scientists to experts from throughout the international metabolomics field. MetaboMeeting 2017 continues to highlight the work of its attendees through both oral platform presentation and poster sessions. The deadline for oral presentation abstracts is 15th July 2017. The deadline for poster abstracts is 1st October 2017. For further information, visit http://metabomeeting2017.thempf.org/. |
| 14-15
Dec 2017 |
Metabolite identification with the Q Exactive and LTQ Orbitrap Venue: Birmingham Metabolomics Training Centre, School of Biosciences, University of Birmingham, Birmingham, UK This two-day course will provide a hands-on approach to teach the latest techniques and tools available to perform metabolite identification. We will apply these tools on the Q Exactive and LTQ Orbitrap mass spectrometry family. The course is targeted towards students and researchers who are actively applying metabolomics. The course will be led by experts in the field and include significant hands-on experience using both the Q Exactive and LTQ Orbitrap instruments to perform:
Bursaries are now available for PhD students funded by the BBSRC, MRC, or NERC. For further information and registration details, please visit http://www.birmingham.ac.uk/facilities/metabolomics-training-centre/courses/metabolite-identification.aspx or contact bmtc@contacts.bham.ac.uk. |
Metabolomics Jobs |
This is a
resource for advertising
positions in metabolomics.
If you have a job you would
like posted in this
newsletter, please email Ian
Forsythe (metabolomics.innovation@gmail.com).
Job postings will be carried
for a maximum of four issues
(eight weeks) unless the
position is filled prior to
that date.
Jobs
Offered
| Job Title | Employer | Location | Posted | Closes | Source |
|---|---|---|---|---|---|
|
Two
PhD positions |
Robert Koch-Institute | Berlin, Germany | 27-Jul-2017 | 7-Aug-2017 |
Robert
Koch-Institute |
|
One
PhD position (two
years at the
Robert
Koch-Institute,
Berlin,
Germany,
and one
year
at ANU,
Canberra,
Australia) |
Robert
Koch-Institute
and ANU |
Berlin, Germany and Canberra, Australia | 27-Jul-2017 | 13-Aug-2017 |
www.allianceberlincanberra.org |
| Seeking qualified applicants for a Postdoctoral Researcher in Developing High-throughput Metabolomics Methods for Populations-based Phenotyping | Karolinska Institutet | Gunma University, Japan | 18-Jul-2017 | 31-Aug-2017 |
Karolinska
Institutet |
| Postdoctoral Researcher in the functional metabolomics of lipid mediators in inflammation | Karolinska Institutet | Solna, Sweden | 6-Jul-2017 | 31-Jul-2017 |
Naturejobs |
| Postdoc Position in Computational Metabolomics | Friedrich Schiller University Jena | Jena, Germany | 4-Jul-2017 | Until filled | Friedrich Schiller University Jena |
| Postdoctoral Fellow / Research Associate in the field of (Bioinformatics / Metabolomics) | McGill University | Montreal, Canada | 25-Jun-2017 | 1-Oct-2017 | McGill University |
| Early
Stage
Researcher (ESR/PhD) |
Biocrates Life Sciences AG | Innsbruck,
Austria |
13-Jun-2017 | Biocrates Life Sciences AG | |
| Analytical
Scientist (PhD or
Master) for Mass
Spectrometry based
Metabolomics |
Biocrates Life Sciences AG | Innsbruck, Austria | 13-Jun-2017 |
|
Biocrates Life Sciences AG |
|
Ian J. Forsythe Double AB, MSc Editor, MetaboNews Department
of Computing
Science
University of Alberta 221 Athabasca Hall Edmonton, AB, T6G 2E8, Canada Email: metabolomics.innovation@gmail.com Website: http://www.metabonews.ca LinkedIn: https://ca.linkedin.com/in/ian-forsythe-465512128 Twitter: http://twitter.com/MetaboNews Google+: https://plus.google.com/118323357793551595134 Facebook: http://www.facebook.com/metabonews |
This
newsletter is
published in
partnership between
The Metabolomics
Innovation Centre
(TMIC, http://www.metabolomicscentre.ca/)
and the Metabolomics
Society (http://www.metabolomicssociety.org)
for the benefit of
the worldwide
metabolomics
community.
|