Workflow4Metabolomics
2.0: New workflows for LC-HRMS, GC-MS, and NMR data
processing, statistical analysis, and annotation
Étienne A. Thévenot1, Marie Tremblay-Franco1,
Misharl Monsoor2, Marion Landi1, Yann
Guitton3, Gildas Le Corguillé2,
Mélanie Pétéra1, Christophe Duperier1,
Jean-François Martin1, Pierrick Roger1,
Franck Giacomoni1, Christophe Caron2
1. MetaboHUB: The
French infrastructure for metabolomics and fluxomics
2. IFB:
French Bioinformatics Institute
3. CNRS IRISA/LINA
contact@workflow4metabolomics.org
What is W4M?
W4M is an online
resource for computational metabolomics built upon the Galaxy
environment (Giacomoni
et al., 2015). W4M currently contains 28 modules
for pre-processing, statistical analysis, and annotation
of data from LC-HRMS, GC-MS, and NMR instruments (Figure
1). The high-performance computing environment of W4M
(600 cores, 100 TB) enables to analyze datasets of more than
several hundreds of samples in a few hours while
requiring only 1% of the resources. Via the homepage users
can open a
private account and access the infrastructure. More than
130 user accounts have been opened in less than one year. W4M
is a collaborative project: it is currently developed
and maintained by a core team of bioinformaticians from the French
Bioinformatics Institute (IFB)
and the French infrastructure for metabolomics and
fluxomics (MetaboHUB;
see the Sep.
2014 MetaboNews Interview).
|
|
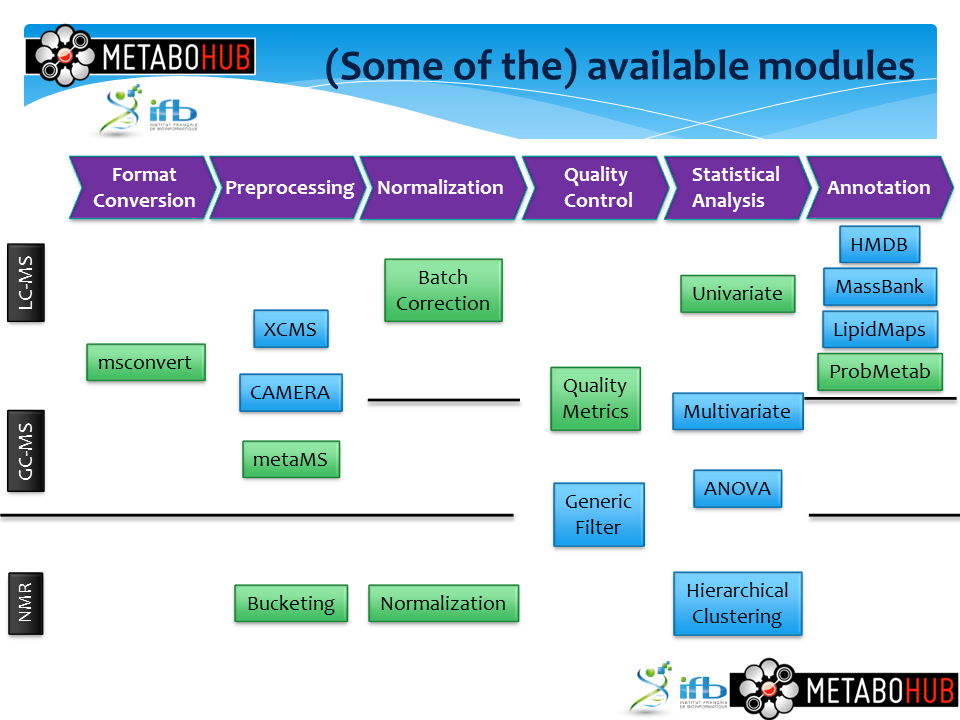
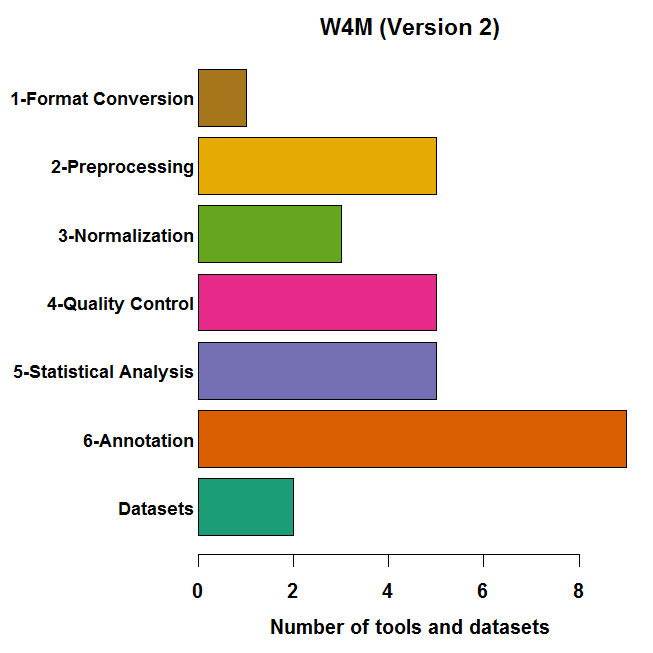
Figure 1. Available modules and datasets in W4M.
Left: The 28 modules cover all the steps from data
conversion and pre-processing, through normalization and
statistics, to annotation. Right: Some modules are
specific to one technology (e.g., for pre-processing and
annotation) while others are common (e.g., for statistical
analysis).
W4M for Users
W4M provides all the tools to build a comprehensive LC-HRMS
workflow starting with format conversion, continuing
with data
processing, followed by normalization
and quality control, performing univariate
and multivariate statistical analyses, and ending with annotation
(Figure
2). Tutorials,
shared workflows, and histories allow beginners to learn how
to conduct their analyzes. Furthermore, the help desk (support@workflow4metabolomics.org)
from the Core Team provides support regarding all
functionalities of W4M.
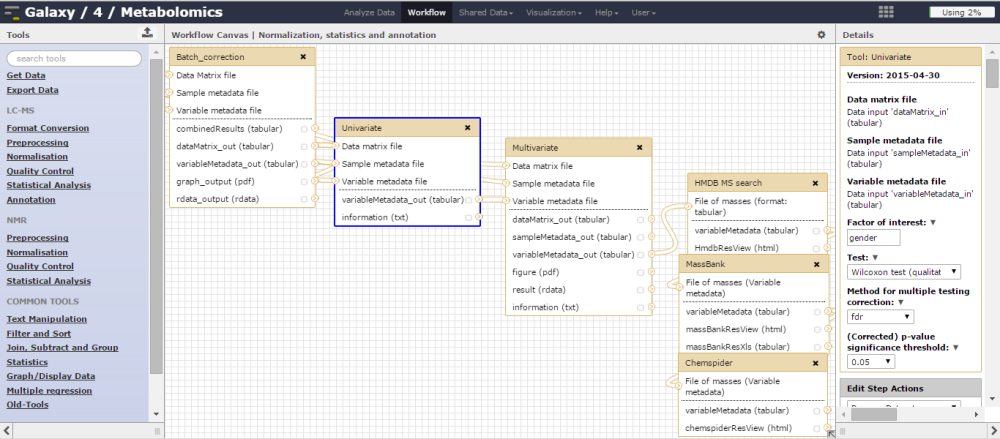 Figure 2.
Figure 2. Creating a workflow for data processing,
statistical analysis, and annotation is straightforward.
W4M for Developers
Building
a Galaxy module from a script (whatever the programming
language and the operating system) is straightforward, requiring
only a configuration file (.xml) and a wrapper to integrate the
tool into the Galaxy interface and handle the inputs/outputs.
W4M 1.0 is available for download as a
virtual
machine. Developers can therefore install the instance
locally and test the integration of their own modules. In
addition, a toolshed will be available soon to share the modules
individually. W4M is therefore a collaborative VRE and new
contributors are welcome.
What’s New in Version 2.0?
The new release (June 1
st, 2015;
Figure
3) provides advanced modules for LC-MS analysis in
addition to new tools for GC-MS and NMR pre-processing.
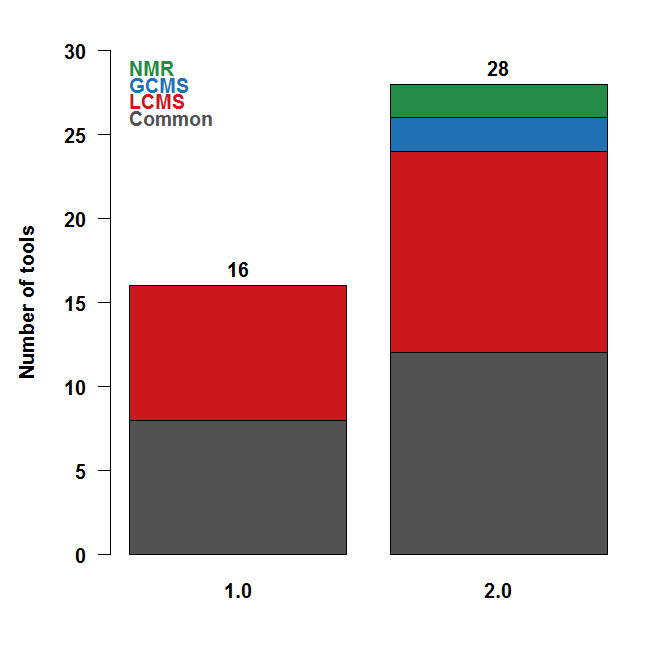
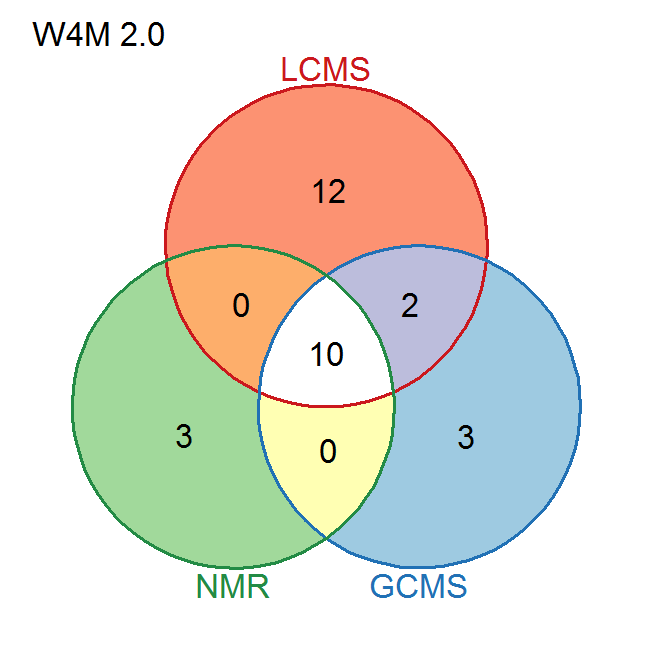
Figure 3. Summary of some of the available
tools for LC-MS, GC-MS, and NMR data analysis.
Left: New
modules (or modules with major updates) are in green.
Right:
The 2.0 release contains advanced functionalities for LC-HRMS,
in addition to specific pre-processing modules for NMR and
GC-MS.
NMR preprocessing modules include bucketing (
Figure
4) and normalization (e.g., Probabilistic Quotient
Normalization).
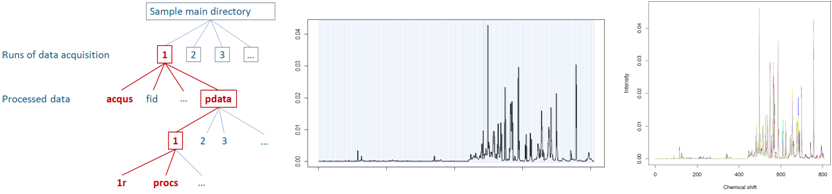 Figure 4.
Figure 4. NMR spectra bucketing and normalization.
Files resulting from upstream pre-processing with the TopSpin
software (
left) can be imported into W4M for bucketing
and integration (
NMR bucketing module;
right),
followed by normalization (
NMR normalization module), and
subsequently analyzed with the statistical modules.
Advanced LC-MS functionalities (24 modules) include
signal drift and batch-effect correction (
Figure
5), quality control of samples and variables (Hotelling's
T2, intensity quantiles), N-way ANOVA, (O)PLS(-DA), and
annotation with in-house or public databases.
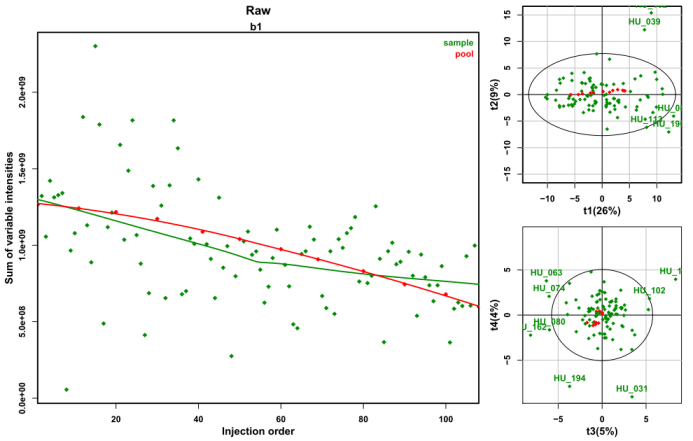
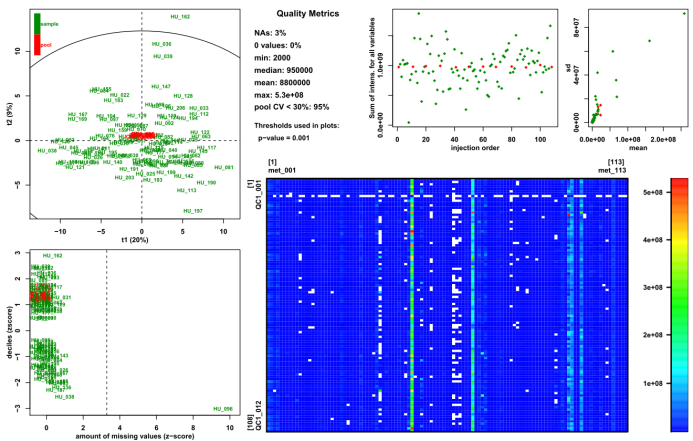
Figure 5. Advanced MS tools for signal drift
correction and quality control.
Left: The signal drift
observed on the raw data can be corrected with the
Batch
Correction module.
Right: The presence of sample
outliers can be then assessed with the
Quality Metrics
module.
What’s Next?
The
W4M
course 2015 (21-25 September 2015, Roscoff, France) will
teach experimenters about W4M features and allow them to
practice with their own data. (Note: for this first session,
presentations will be in French but slides will be in English.)
W4M will continue to be enriched with new modules. Future
development will include new functionalities for MS
2
analysis and NMR annotation, in addition to pathway annotation
through a connection to
MetExplore,
and raw data export to the
MetaboLights
repository.
Acknowledgment: W4M is supported by the grants from
MetaboHUB [ANR-11-INBS-0010], IFB [ANR-11-INBS-0013],
Biogenouest®, Lifegrid (Auvergne), and by the IDEALG project
[ANR-10-BTBR-04].
Please note: If you know of any
metabolomics research programs, software, databases,
statistical methods, meetings, workshops, or training
sessions that we should feature in future issues of this
newsletter, please email Ian Forsythe at metabolomics.innovation@gmail.com.