
Issue 26 - October 2013
CONTENTS:
Online version of this newsletter:
http://www.metabonews.ca/Oct2013/MetaboNews_Oct2013.htm
 |
| A
newsletter published in partnership between TMIC and
the Metabolomics Society Issue 26 - October 2013 |
|
CONTENTS: |
|
|
Press Release: http://www.metabonews.ca/Oct2013/press_releases/TMIC-Met_Soc_Partnership_Press_Release.htm
 |
Metabolomics Society News |
| |
Platform Spotlight
|
 |
 |
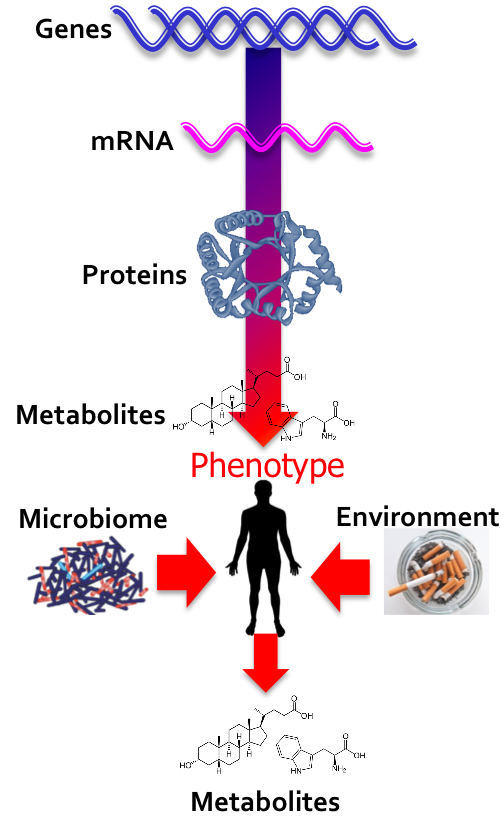
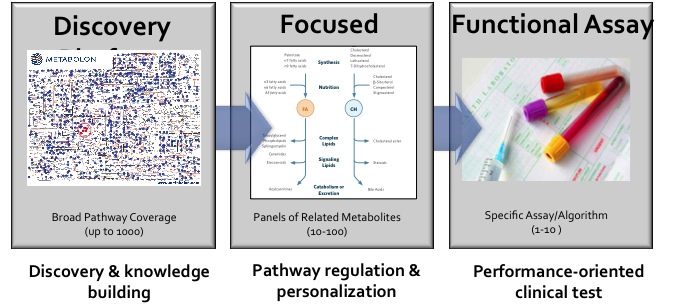
| |
MetaboInterviews
|
|
|
Professor,
Chemistry and Integrative Genomics, Princeton
University, Princeton, NJ, USA |
 |
Biography Joshua Rabinowitz
grew up in Chapel Hill, North Carolina. In 1994, he
earned B.A. degrees in Mathematics and Chemistry from
the University of North Carolina. From there, he moved
west to Stanford, where he earned his Ph.D. in
Biophysics in 1999, followed by his M.D. in 2001. In
2000, he co-founded Alexza Pharmaceuticals leading
R&D efforts there for four years. In 2004, Joshua
returned to academia, joining the faculty of Princeton
University where he is currently Professor of Chemistry
and Integrative Genomics. His lab focuses on
understanding cellular metabolism, its normal
regulation, and its dysregulation in disease. He is the
author of more than 100 papers and the inventor of over
100 patents that include the FDA-approved Adasuve
inhaler.
http://www.princeton.edu/genomics/rabinowitz/research/ |
| |
Biomarker Beacon
|
 |
Metabolomics Current
Contents
|
 |
MetaboNews |
| 27 Sep 2013 |
The Global
Metabolomics Market 2013 Report: An In-Depth
Analysis and Forecast of the Market Covering
the Leading Competitors in Metabolomics
Research and Markets (http://www.researchandmarkets.com/research/hg72tl/metabolomics) have announced the addition of the "The Global Metabolomics Market 2013 Report" report to their offering. Over the last decade, genomics and proteomics have been used as key tools to discover potential drug targets and to better understand the complexities of biology. To balance research in these areas, metabolomics, a new science is evolving for analyzing the basic metabolic changes taking place in a living organism. Metabolomics is thus an emerging concept which refers to the systematic study of the distinctive chemical fingerprints generated in a particular cellular process. This report covers the market by disease indications along with the applications and metabolite profiling for biomarker discovery applications. In addition, it also includes the factors driving and restraining the market and covers the market scenario in the U.S., Europe, Asia and the Rest of the World (ROW). This report will provide the company profiles of key companies along with the competitive analysis. The Global Metabolomics market is showing a double digit growth (CAGR 35%) due to supportive factors such as: (i) increased willingness of biotechnology and pharma companies to adopt metabolomics concept to drive R&D activity within the industry, (ii) rapid growth of metabolomics data analysis softwares and solutions, and (iii) the advancement of analytical technologies. Metabolomics is used in the identification of new biomarkers, which indicate a change in the physiological state of a cell or tissue. Biomarker screening is important in the process of new drug discovery, and is also a key in vitro diagnostics tool. Additionally it is also being used for environmental toxicology screening. While, there are major concerns of this market such as the validation challenges, the positive aspects may very well offset the market restraints to aid the market grow at an exceptional rate. |
| 20 Sep 2013 |
NIH Funds Three
Metabolomics Resource Centers
The National Institutes of Health has provided tens of millions of dollars in new funding to support three new Comprehensive Metabolomics Centers at Mayo Clinic, the University of Kentucky, and the University of Florida that will support metabolomics research in their regions. These three centers will receive an estimated $9 million to $10 million each over the next five years to create resources and initiate research programs that will ramp up the national metabolomics science capabilities. Funded by the NIH Common Fund and Coordinated by the National Institute of Diabetes and Digestive and Kidney Diseases, these centers in Minnesota, Kentucky, and Florida will serve as metabolomics hubs and resources for research around the country. They will provide a range of resources and training programs, and will support a variety of initiatives, including providing metabolomics-related services, such as biomarker identification, bioinformatics analyses, mass spectrometry, and nuclear magnetic resonance and other imaging services. Last year, NIH provided $14.3 million to fund the launch of three other metabolomics resource centers at the University of Michigan, the Research Triangle Institute, and the University of California, Davis. |
| 18 Sep 2013 |
Mayo Clinic
Receives $8.8 Million Federal Grant for
Metabolomics Center
Mayo Clinic is one of six new federally-funded Comprehensive Metabolomics (met-ah-bol-OH-mics) Centers to support medical research on metabolomics -- the study, at the cellular level, of how molecules are metabolized in the body. The award from the National Institutes of Health (NIH) is for $8.8 million over five years. “It is certainly an honor to be selected as one of the six national centers which will serve as resources for universities and other research institutions. This offers a tremendous opportunity for Mayo Clinic to be a national leader in this emerging research area,” says K. Sreekumaran Nair, M.D., Ph.D., Mayo Clinic endocrinologist and principal investigator on the grant, as well as head of Mayo’s Comprehensive Metabolomics Core facility. Metabolomics seeks to understand metabolites -- the byproducts of cellular metabolism. These molecules are important because they offer a fingerprint for researchers of the activity going on in the cells of various tissues. They can help understand how disease develops and spreads and they can help identify biomarkers, signs within the body that indicate the beginning of a problem or how well a patient may be responding to treatment for a condition. Metabolomic analysis depends on measuring thousands of small molecules in fluids, such as blood, urine, spinal fluid or fluid inside of cells. The award announcement says the purpose of the centers is to “facilitate institutional development of pioneering research, metabolomics training and outreach programs in this emerging area.” Under the grant, the Mayo Clinic center will offer mass spectrometry and nuclear magnetic resonance spectroscopy for metabolic analysis and will offer training courses both to investigators at Mayo and other institutions. The center also will advise on study design and application of stable isotopes, especially based on mass spectrometry. Other Mayo Clinic co-investigators on the grant are Adrian Vella, M.D., Michael Joyner, M.D., Michael Jensen, M.D., Petras Dzeja, Ph.D., and Slobodan Macura, Ph.D. Technical staff at the center are led by G. Charles Ford and Mai Persson. The grant (U24DK100469) is coordinated by the National Institute of Diabetes and Digestive and Kidney Diseases, but originates from the NIH Director’s Common Fund. |
 |
Metabolomics Events
|
| 1-3 Oct 2013 |
The 10th International
Symposium on Milk Genomics and Human Health The venue for this year's event is the U.C. Davis Conference Center located on the University of California, Davis campus in the United States. Program The three day event will bring together international experts in nutrition, genomics, bioinformatics and milk research to discuss and share the latest breakthroughs and their implications. The Annual Symposium is our flagship event that features scientific research related to milk and human health done throughout the world. The symposium draws from the diversity of its memberships to cover the breath of genomics themes that reflect the interest of the Consortium. The goal of the Consortium is to bring together the research and dairy communities to share, translate, and interpret data that are happening within the fields of the "-omics" science. For more information, visit http://milkgenomics.org/10th-international-symposium-on-milk-genomics-and-human-health/. |
| 7-11 Oct 2013 |
Metabolomics course:
SLC-Tjärnö marinebiological laboratory Application should include a short motivation (<1 page) and a brief CV. Submit by E-mail to erik.selander@bioenv.gu.se Application deadline 15th of August 2013 Teachers: Prof. Georg Pohnert, Biorganic Analytics, Friedrich Schiller University, Jena Prof. Johan Trygg, Department of Chemistry, Umeå University Dr. Ulf Sommer, NERC Metabolomics Facility, University of Birmingham Contact and inquiries: Erik Selander erik.selander@bioenv.gu.se Göran Nylund goran.nylund@bioenv.gu.se Course Flyer: http://www.metabonews.ca/May2013/events/Metabolomics%20course%20flyer.pdf |
| 7-11 Oct 2013 |
Hands-on LC-MS for
Metabolic Profiling Course Day 1 Introductory lectures in mass spectrometry and chromatography, study design and sample preparation. Days 2 & 3 Analysis of biofluids through global profiling and targeted analyses; one day spent on each of the newest QToF instrumentation and the newest TQ instrumentation. Instrument set up, method development and acquisition will be covered. As we have set a maximum of 4 attendees per instrument this allows for hands-on participation by all. Day 4 Lectures in data analysis, followed by workshops where attendees will process the data acquired from the previous day, allowing for development of interpretation skills. Day 5 Application lectures, tips, tricks and troubleshooting. Download the full programme here: LCMS_Metabolic_Profiling For more information, visit http://www1.imperial.ac.uk/iiptc/courseinfo/lcms/. |
| 4 Nov 2013 |
Venomics: Drug Discovery
from Nature's Deadliest |
| 17-21 Mar 2014 |
EMBO Practical Course on
Metabolomics Bioinformatics for Life
Scientists This course will provide an overview of key issues that affect metabolomics studies, bioinformatics tools, and procedures for the analysis of metabolomics data. It will be delivered using a mixture of lectures, computer-based practical sessions and interactive discussions. The course will provide a platform for discussion of the key questions and challenges in the field of metabolomics. Audience This course is aimed at PhD students and researchers with a minimum of one year’ s experience in the field of metabolomics who are seeking to improve their skills in metabolomics data analysis. Participants must have experience using R (including a basic understanding of the syntax and ability to manipulate objects) and the UNIX/LINUX operating system. For more information, visit http://www.ebi.ac.uk/training/course/metabolomics-2014. |
| 24-26 Mar 2014 |
3rd International
Conference and Exhibition on Metabolomics
& Systems Biology The annual Metabolomics conference mainly aims in bringing Metabolomics and Systems Biology researchers from around the world under a single roof, where they discuss the research, achievements and advancements in the field. After the success of Metabolomics-2012 & Metabolomics-2013, OMICS Group is proud to announce the 3rd International Conference and Exhibition on Metabolomics & Systems Biology which is going to be held during March 24-26, 2014 at Hilton San Antonio Airport, USA. Metabolomics-2014 meeting promises a program full of practical workshops and parallel sessions covering the broad range of biological and technological metabolomics topics, providing rich opportunities for networking and approach towards biomedical and biological scientific research. Join us at Metabolomics-2014 as we gather together to share ideas, insights and advances in the field of Metabolomics and Systems Biology. Conference Highlights
For more information, visit http://www.metabolomicsconference.com/. |
| 23-26 Jun 2014 |
Metabolomics 2014: 10th
Annual International Conference of the
Metabolomics Society Early registration and abstract submission due March 31, 2014. Follow us on Twitter:
Please come back later for detailed information about Metabolomics 2014 by visiting http://metabolomics2014.org. |
 |
Metabolomics Jobs |
This is a resource for
advertising positions in metabolomics. If you have a job
you would like posted in this newsletter, please email
Ian Forsythe (metabolomics.innovation@gmail.com).
Job postings will be carried for a maximum of 4
issues (8 weeks) unless the position is filled prior to
that date.
Jobs
Offered
| Job Title | Employer | Location | Posted | Closes | Source |
|---|---|---|---|---|---|
| An engineer-technician in metabolomics | CRP - Gabriel Lippmann | Belvaux, Luxembourg | 23-Jul-2013 | 31-Dec-2013 |
Metabolomics
Society |
| Program Coordinator | University of Florida |
Gainesville,
USA |
26-Sep-2013 | 17-Nov-2013 |
University
of Florida |
| Postdoctoral Researcher in Metabolomics of Pulmonary Medicine | Karolinska Institutet | Stockholm, Sweden | 15-Sep-2013 | 15-Nov-2013 |
naturejobs.com |
| National Research Council (NRC) post-doctoral fellowship positions | Environmental Protection Agency (EPA) |
Athens,
USA |
30-Sep-2013 | 1-Nov-2013 |
EPA |
| Senior Research Scientist - NMR Metabolomics | Department of Primary Industries, Victoria |
Bundoora, Australia |
10-Sep-2013 | 31-Oct-2013 |
naturejobs.com |
| Postdoctoral Position in Metabolic Studies of Cancer Models | University of California San Francisco |
San
Francisco, USA |
14-Sep-2013 | Metabolomics Society | |
| Canada Research Chair (Tier II) in Marine Microbial Proteomics and Metabolomics | Dalhousie University |
Halifax,
Canada |
20-Aug-2013 | 15-Oct-2013 |
naturejobs.com |
| AIHS Translational Health Chair - Metabolomics, Department of Biological Sciences, Faculty of Science | University of Calgary | Calgary, Canada | 11-Aug-2013 | 8-Oct-2013 |
naturejobs.com |
|
Ian
J. Forsythe, M.Sc.
MetaboNewsEditor Department of Computing Science
University of Alberta 221 Athabasca Hall Edmonton, AB, T6G 2E8, Canada Email: metabolomics.innovation@gmail.com Website: http://www.metabonews.ca LinkedIn: http://ca.linkedin.com/in/iforsythe Twitter: http://twitter.com/MetaboNews Google+: https://plus.google.com/118323357793551595134 Facebook: http://www.facebook.com/metabonews |
This newsletter is
published in partnership between The
Metabolomics Innovation Centre (TMIC, http://www.metabolomicscentre.ca/)
and the Metabolomics Society (http://www.metabolomicssociety.org)
for the benefit of the worldwide
metabolomics community.
 A single source destination for fee-for-service metabolic profiling including comprehensive metabolite identification, quantification, and analysis 
|