
TMIC and the Metabolomics Society
Issue 38 - October 2014
CONTENTS:
Online version of this newsletter:
http://www.metabonews.ca/Oct2014/MetaboNews_Oct2014.htm
 |
| Published
in partnership between TMIC and the Metabolomics Society Issue 38 - October 2014 |
|
CONTENTS: |
|
|
 |
Metabolomics Society News |

| |
Metabolomics Spotlight
|
| |
MetaboInterviews
|
|
|
Assistant Professor of Applied Biostatistics, Department of Medicine, University of Alberta |
 |
Biography Dr.
David Broadhurst, born in Chester, UK, holds a first
class honors degree in Electronic Engineering, an MSc in
Medical Informatics, and a PhD in the "Application of
Artificial Neural Networks and Evolutionary Algorithms
to Metabolic Profiling". He has been an active member of
the metabolomics community for the last 18 years, where
he is a recognized expert in design of experiments,
signal processing, biostatistics, and machine learning.
David worked for an extended period as a post-doctoral
research fellow developing large-scale clinical
metabolomics protocols alongside Dr. Warwick Dunn, at
the University of Manchester as part of Professor
Douglas Kell’s Bioanalytical Sciences Group. In 2009, he
moved to Cork University Maternity Hospital, Ireland, to
investigate pre-symptomatic metabolite biomarkers
predictive of major pregnancy diseases. In January 2011,
David took up his current position as Assistant
Professor of Applied Biostatistics in the department of
Medicine, at the University of Alberta, Canada, where he
is scientific lead and academic coordinator for a range
of clinical metabolomics, and integrated polyomic,
personalized medicine projects. His current research
primarily focuses on developing validated parsimonious
prediction algorithms, combining novel feature selection
algorithms, precise experimental design, and robust
quality assurance procedures.
|
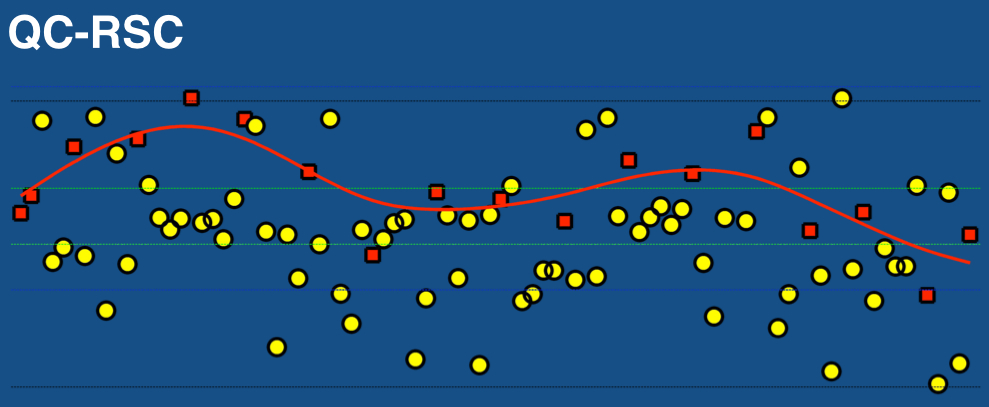
Biomarker Beacon
|
Cobb J, Eckhart A, Perichon R, Wulff J, Mitchell M, Adam KP, Wolfert R, Button E, Lawton K, Elverson R, Carr B, Sinnott M, Ferrannini E. A Novel Test for IGT Utilizing Metabolite Markers of Glucose Tolerance. J Diabetes Sci Technol. 2014 Sep 26. pii: 1932296814553622. [Epub ahead of print] [PMID: 25261439]
In recent years, health care practitioners rely decreasingly on the oral glucose tolerance test (OGTT) for the diagnosis of patients with impaired glucose tolerance (IGT). In recent studies, researchers used metabolic profiling to identify a number of metabolites that are associated with dysglycemia and type 2 diabetes. In this paper, the research team used quantitative assays for 23 candidate biomarker metabolites and statistical analysis to compare fasting plasma samples from patients with IGT and normal glucose tolerance (NGT). They developed a model based on a set of metabolites (glucose, α-hydroxybutyric acid, β-hydroxybutyric acid, 4-methyl-2-oxopentanoic acid, linoleoylglycerophosphocholine, oleic acid, serine, and vitamin B5) to differentiate between IGT and NGT patients. This metabolite-based test may prove to be superior to OGTT and eventually replace it as a means of identifying patients with IGT.
 |
Metabolomics Current
Contents
|
This section of
MetaboNews is supported by:
|
Metabolomics Events
|
| 23-24 Oct 2014 |
Challenges and advances in
the annotation and de novo identification of
small molecules of biological origin For more details, please visit http://www.icsn.cnrs-gif.fr/spip.php?rubrique191&lang=en. |
| 28-31 Oct 2014 |
Metabolomics Beginners
Workshop MA at the University of Melbourne (UM) along with the Australia and New Zealand Metabolomics Network (ANZMN) is running a 4-day beginners workshop on metabolomics. This workshop focuses on an analytical component (Tuesday 28th October and Wednesday 29th October) and data processing and statistical analysis of metabolomics data (Thursday 30th October and Friday 31st October) obtained through GC-MS and LC-MS analytical platforms. The workshop is open to all researchers from academic, government and commercial organisations. This workshop would assist researchers working in the area of metabolomics, as well as researchers planning on using metabolomics in any future studies. Travel awards: 3x Metabolomics Society $US 500 travel awards for interstate/international student attendees *Students must be members of the Metabolomics Society *Awards will be based on a 200-word abstract of the student's research project & a two page CV Cost and Registration: http://ecommerce.bio21.unimelb.edu.au/categories.asp?cID=15 For more details, please visit https://sites.google.com/site/anzmnworkshop2014/home |
| 29-30 Oct 2014 |
Clinical Applications of
Mass Spectrometry As the analytical power of mass spec is realised, the range of applications using this technology continues to expand. Focus at this meeting will be given to both traditional & emerging uses of MS in the clinic. Hot topics to be covered include developments of MS in applications ranging from vitamin D detection to newborn blood spot analysis. Attending this event will provide you with excellent opportunities for networking with like minded peers, helping you to build new relationships and optimise your workflow. Running alongside the conference will be an exhibition covering the latest technological advances and associated services from leading solution providers within this field. Registered delegates will also have access to the co-located Food Analysis Congress, ensuring a cost effective trip. Keynote Speakers:
For more details, please visit http://selectbiosciences.com/conferences/index.aspx?conf=CAMS2014 |
| 29-30 Oct 2014 |
Food Analysis Congress Focus will be given to advances in both the analysis of natural food allergens and toxins, as well as contaminants introduced through processing and packaging. Points for discussion will also include the ongoing issue of food traceability and efforts to reduce food fraud. Attending this event will provide you with excellent opportunities for networking with like minded peers, helping you to find solutions and build collaborations. Running alongside the conference will be an exhibition covering the latest technological advances and associated services from leading solution providers within this field. Registered delegates will also have access to the co-located Clinical Applications of Mass Spectrometry track, ensuring a cost effective trip. For more details, please visit http://selectbiosciences.com/conferences/index.aspx?conf=FAC2014 |
| 10-11 Nov 2014 |
Metabolomic Approaches:
Advanced Analytical Tools Anisha Wijeyesekera (London Imperial College, United Kingdom) Oscar Yanes (Centre for Omic Sciences, Spain) Ron Wehrens (Wageningen UR, The Netherlands) Matteo Stocchero (S-IN Soluzioni Informatiche, ltaly) Download the Event Flyer For further details, please visit http://cittadellasperanza.org/metabolomic-approaches/. |
| 13 Nov 2014 |
5th Danish Symposium on
Metabolomics Metabolomics concerns the comprehensive characterization of small molecule metabolites in biological systems. It can provide an overview of the metabolic status and global biochemical events associated with a cellular or biological system. The main topic of the 5th Danish Symposium on Metabolomics is metabolomics research in Denmark; peak annotation and identification. We encourage researcher from Danish institutions to submit an abstract for oral presentation of 15 min duration. As last year we will include a poster session and we urge participants to bring posters with metabolomics related research. Oral presentation and poster abstracts should be submitted to jch@plen.ku.dk no later than 27th October. For further details, please consult the event flyer. |
| 14 Nov 2014 |
1st Nordic Symposium on
Multidimensional Chromatography The peak capacity of one-dimensional chromatographic systems such as LC, GC, CE and SFC is often insufficient to provide baseline separation of compounds in complex mixtures such as in environmental, biological, petrochemical, and food samples, but also in the pharmaceutical industry one-dimensional systems are often not able to provide sufficient separation of impurities. Multidimensional chromatography allows separation of complex mixtures by using multiple columns with different stationary phases. These columns are coupled with an interface that allows the fractions from the first column to be selectively transferred to other columns for additional separation. The main topics of the 1st Nordic Symposium on Multidimensional Chromatography cover the fundamentals of comprehensive multidimensional chromatography, where all fractions from the first (or second) column are transferred to the second (or third) column for additional separation. Four international experts will give 6 hours of lectures. For further details, please consult the event flyer. |
| 17-18 Nov 2014 |
Waters Metabolic Profiling
Deminar in collaboration with Imperial College
London On day one attendees will meet at Heathrow for lunch and an introduction to the collaboration between Imperial College London & Waters. Guests will be transferred to the MRC NIHR National Phenome Centre where they will have the opportunity for a guided tour to see large scale metabolic phenotyping in action. Day two will take place at the Imperial International Phenome Training Centre (IIPTC), Imperial College South Kensington campus, where participants will listen to guest speakers from Imperial College London and interact with both Imperial & Waters scientists who run the state of the art instrumentation through a series of planned interactive laboratory workshops. In addition, registrants are encouraged to stay on for a few more days and participate in the “Metabolic Phenotyping in Disease Diagnosis & Personalised Health Care” course hosted by Imperial College London at the Imperial International Phenome Training Centre. This course offers seminars in the techniques and applications of LC-MS, GC-MS, MS imaging and nuclear magnetic resonance (NMR) spectroscopy in the field of metabolic profiling, as well as metabolite identification and multivariate statistics. This is complemented by hands-on workshops focusing on data analysis using statistical techniques such as PCA and O2-PLSDA. Learn more on Waters metabolomics and lipidomics. For further details, please visit the event website. |
| 18-21 Nov 2014 |
Workshop in Metabolic
Phenotyping in Disease Diagnosis &
Personalised Health Care Download the full programme here: Metabolic Profiling Disease Diagnosis For further details, please visit the workshop website. |
| 19-20 Nov 2014 |
Separation Science Asia
2014 You will gain valuable practical, technical and solutions-based knowledge for getting the most out of your HPLC, GC, MS and sample preparation methods, improving your skills, knowledge and job productivity. In addition, you will be able to interact directly with these technology leaders at the conference: Click here for more details on the programme. For further details, please visit the conference website. |
| 19-21 Nov 2014 |
Merlion Metabolomics
Workshop Singapore 2014 This 3-day workshop features three important areas of metabolomics research, namely, Environment, Food Science and Technology, and Human Health. Eight tracks, including two sessions for young researchers to share their research findings, will facilitate knowledge sharing and discussions over 2.5 days during the workshop. A half day technical site visit to research and/or manufacturing facilities will be arranged for a selected group of participants. Prominent scientists in metabolomics from Asia-Pacific region will also be invited to the workshop. For further details, please visit the workshop website. |
| 3-5 Dec 2014 |
2nd
Australian Lipid Meeting While the first meeting focused on lipidomics we have expanded the scope for the second meeting to cover all aspects of lipid research. Planned topics include:
Registration
|
| 11-13 Dec 2014 |
2nd ICAN (Institute of
Cardiometabolism and Nutrition) Conference
Series 2014 Further Information: http://ican-series.com/ For further details, visit the conference
website. |
| 28 Jun to 2 Jul 2015 |
Metabolomics 2015: 11th
Annual International Conference of the
Metabolomics Society For further details, visit http://metabolomics2015.org. |
Metabolomics Jobs |
This is a resource for
advertising positions in metabolomics. If you have a job
you would like posted in this newsletter, please email
Ian Forsythe (metabolomics.innovation@gmail.com).
Job postings will be carried for a maximum of 4
issues (8 weeks) unless the position is filled prior to
that date.
Jobs
Offered
| Job Title | Employer | Location | Posted | Closes | Source |
|---|---|---|---|---|---|
| Research
Officer (Metabolomics Australia) |
University of
Melbourne |
Melbourne, Australia | 1-Oct-2014 |
22-Oct-2014 |
The Conversation Job Board |
| Post
Doctorate RA - Metabolomics |
Pacific Northwest National Laboratory | Richland, WA, United States | 24-Sep-2014 |
18-Oct-2014 |
Metabolomics Society Jobs |
| Post
Doctorate - Cheminformatics |
Pacific Northwest National Laboratory | Richland, WA, United States | 24-Sep-2014 |
15-Oct-2014 |
Metabolomics Society Jobs |
| Research
Fellow - Metabolomics |
Mayo Clinic |
Rochester, MN, United States | 15-Sep-2014 |
Mayo Clinic | |
| Research
Engineer in Metabolomics |
The Roullier Group | Dinard, France | 11-Sep-2014 |
Réseau Français de Métabolomique et Fluxomique | |
| Post-Doc
Subcellular fluxomics in plant cells |
MetaboHUB |
Bordeaux, France | 11-Sep-2014 |
30-Oct-2014 |
MetaboHUB |
| Research
Programmer |
University of Alberta | Edmonton, Canada | 2-Sep-2014 |
University of Alberta | |
| PhD
Studentship |
International
Pregnancy Research Alliance (IPRA) |
Chongqing, China | 27-Aug-2014 |
31-Dec-2014 |
Metabolomics Society Jobs |
| Mass Spectrometry Technician | International Pregnancy Research Alliance (IPRA) | Chongqing, China | 27-Aug-2014 | 31-Dec-2014 | Metabolomics Society Jobs |
| Analytical
Scientist
(Research & Development) |
BIOCRATES Life Sciences AG |
Innsbruck, Austria |
1-Aug-2014 |
BIOCRATES Life Sciences AG | |
| Business
Development Manager USA |
BIOCRATES Life Sciences AG | Innsbruck, Austria | 1-Aug-2014 | BIOCRATES Life Sciences AG |
Equipment
|
Ian
J. Forsythe, M.Sc.
MetaboNewsEditor Department of Computing Science
University of Alberta 221 Athabasca Hall Edmonton, AB, T6G 2E8, Canada Email: metabolomics.innovation@gmail.com Website: http://www.metabonews.ca LinkedIn: http://ca.linkedin.com/in/iforsythe Twitter: http://twitter.com/MetaboNews Google+: https://plus.google.com/118323357793551595134 Facebook: http://www.facebook.com/metabonews |
This newsletter is
published in partnership between The
Metabolomics Innovation Centre (TMIC, http://www.metabolomicscentre.ca/)
and the Metabolomics Society (http://www.metabolomicssociety.org)
for the benefit of the worldwide
metabolomics community.
 A single source destination for fee-for-service metabolic profiling including comprehensive metabolite identification, quantification, and analysis 
|