
TMIC and the Metabolomics Society
Issue 47 - July 2015
CONTENTS:
Online version of this newsletter:
http://www.metabonews.ca/Jul2015/MetaboNews_Jul2015.htm
 |
| Published
in partnership between TMIC and the Metabolomics Society Issue 47 - July 2015 |
|
CONTENTS: |
|
|
 |
Metabolomics Society News |
| |
Metabolomics Spotlight
|
 |

| |
MetaboInterview
|
| Senior
Lecturer in Analytical Chemistry, RMIT University, Melbourne, Australia |
|
 |
Biography Dr Oliver (Oli) Jones is a senior lecturer in analytical chemistry at RMIT University in Melbourne, Australia. He obtained his PhD from Imperial College London in 2005, then worked as a postdoc in the department of Biochemistry at the University of Cambridge, and later as a lecturer in the School of Engineering and Computing Sciences at the University of Durham. Oli is currently president of Proteomics and Metabolomics Victoria and the Australian and New Zealand Metabolomics Network (for whom he writes a weekly newsletter); secretary of the Australian and New Zealand Society for Magnetic Resonance; a CI at the Australian Centre for Research on Separation Science and a member the Australian Academy of Science, National Committee for Chemistry. Aside from metabolomics, his group also conducts research in analytical methods and technologies, particularly in multidimensional chromatographic techniques, for a range of applications including environmental food and forensic chemistry. He is also deputy director of the RMIT Centre for Environmental Sustainability and Remediation and serves on the editorial board of the Journal of Environmental Immunology and Toxicology. He has 55 publications with a total of over 2300 citations and an h-index of 19. |
 OJ: Recently my
group has focussed more on the analytical chemistry side of
metabolomics, particularly separation science. We have been
applying online two-dimension liquid chromatography (2DLC)
methods to increase the number of metabolites we can pick up
in single run for example (see MetaboNews January
2015—http://www.metabonews.ca/Jan2015/MetaboNews_Jan2015.htm#spotlight).
So far we have applied 2DLC to the untargeted analysis of
mushrooms and the targeted analysis of cannabinoids in hemp.
This work won an outstanding investigator award at the
Metabolomics Society meeting in Japan last year—so hopefully I
am doing something right. My group has recently become part of
the Australian Centre for Research on Separation Science
(ACROSS), which was a real honour and has been very
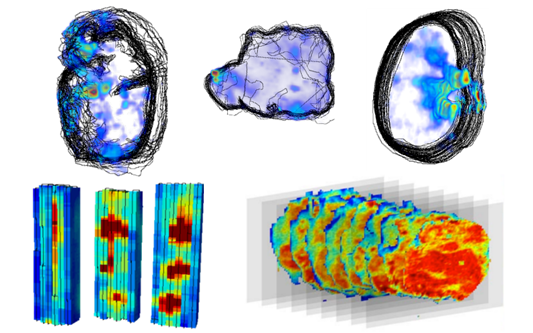
productive. My most recent project is based on using some
highly fluorescent compounds (see image), originally developed
for use in solar cells as tags for the analysis of fatty acids
at low concentrations via HPLC, rather than the more common
FAME analysis via GC-FID.
OJ: Recently my
group has focussed more on the analytical chemistry side of
metabolomics, particularly separation science. We have been
applying online two-dimension liquid chromatography (2DLC)
methods to increase the number of metabolites we can pick up
in single run for example (see MetaboNews January
2015—http://www.metabonews.ca/Jan2015/MetaboNews_Jan2015.htm#spotlight).
So far we have applied 2DLC to the untargeted analysis of
mushrooms and the targeted analysis of cannabinoids in hemp.
This work won an outstanding investigator award at the
Metabolomics Society meeting in Japan last year—so hopefully I
am doing something right. My group has recently become part of
the Australian Centre for Research on Separation Science
(ACROSS), which was a real honour and has been very
productive. My most recent project is based on using some
highly fluorescent compounds (see image), originally developed
for use in solar cells as tags for the analysis of fatty acids
at low concentrations via HPLC, rather than the more common
FAME analysis via GC-FID. OJ: Although I was
born and bred in the UK, I now call Australia home and there
is a lot of great work going on here. Metabolomics Australia
has been going for some time now and is well known. In
Victoria, we also have Proteomics and Metabolomics Victoria
and, since 2012, the Australian and New Zealand Metabolomics
Network (ANZMN), which recently became an official affiliate
of the International Metabolomics Society. We publish a weekly
metabolomics e-newsletter and the team has helped organise a
series of well-respected (and well-attended) workshops and
training sessions. We also get a lot of support from most of
the major instrument vendors, who all see metabolomics as a
growth area so I think the field will continue to grow in
Australia for a while yet.
OJ: Although I was
born and bred in the UK, I now call Australia home and there
is a lot of great work going on here. Metabolomics Australia
has been going for some time now and is well known. In
Victoria, we also have Proteomics and Metabolomics Victoria
and, since 2012, the Australian and New Zealand Metabolomics
Network (ANZMN), which recently became an official affiliate
of the International Metabolomics Society. We publish a weekly
metabolomics e-newsletter and the team has helped organise a
series of well-respected (and well-attended) workshops and
training sessions. We also get a lot of support from most of
the major instrument vendors, who all see metabolomics as a
growth area so I think the field will continue to grow in
Australia for a while yet. OJ: In the short term,
I very much hope to see more applications of multidimensional
chromatography in metabolomics to increase the number of
metabolites we can see in a single run. In the longer term, I
think metabolomics could make a real difference to risk
assessment. For example, the work we did on mixture effects in
environmental toxicology showed that this issue should be
considered in chemical risk assessment (and that metabolomics
could help assess such mixture effects); a later project at
Cambridge, funded by the UK Food Standards Agency, helped
improve our understanding of the risks to humans of exposure to
mixtures of pesticides in food.
OJ: In the short term,
I very much hope to see more applications of multidimensional
chromatography in metabolomics to increase the number of
metabolites we can see in a single run. In the longer term, I
think metabolomics could make a real difference to risk
assessment. For example, the work we did on mixture effects in
environmental toxicology showed that this issue should be
considered in chemical risk assessment (and that metabolomics
could help assess such mixture effects); a later project at
Cambridge, funded by the UK Food Standards Agency, helped
improve our understanding of the risks to humans of exposure to
mixtures of pesticides in food. OJ: Well, given the
sheer number of papers published on metabolomics today, I
think one could argue that the field has already 'taken off'.
Nevertheless, one thing that I think will really drive
advances will be the miniaturisation of analytical
instrumentation. For instance back in the 1980s mass
spectrometers were the size of a small room and required a
computer almost as big, as well as highly trained staff to run
them, and, as such, were the preserve of the few. Once mass
specs were small and robust enough to be used routinely in the
lab, however, the use and applications of mass spectrometry
expanded rapidly. The new wave of benchtop NMR spectrometers
is a great step in the direction of smaller instruments, and
applications of this technique have already started to take
off but I hope this is just the start. Imagine if you had an
LC the size of a computer chip or lab slide (already being
worked on at ACROSS), and/or a mass spectrometer robust enough
for you to transport in (and perhaps even power from) your
car, and as easy to operate as your smartphone? The potential
this would have for metabolomic analysis of samples from huge
numbers of people could drive some incredible work in the
health and medical sciences as well as a range of other
fields. It would be nice to see that in my lifetime.
OJ: Well, given the
sheer number of papers published on metabolomics today, I
think one could argue that the field has already 'taken off'.
Nevertheless, one thing that I think will really drive
advances will be the miniaturisation of analytical
instrumentation. For instance back in the 1980s mass
spectrometers were the size of a small room and required a
computer almost as big, as well as highly trained staff to run
them, and, as such, were the preserve of the few. Once mass
specs were small and robust enough to be used routinely in the
lab, however, the use and applications of mass spectrometry
expanded rapidly. The new wave of benchtop NMR spectrometers
is a great step in the direction of smaller instruments, and
applications of this technique have already started to take
off but I hope this is just the start. Imagine if you had an
LC the size of a computer chip or lab slide (already being
worked on at ACROSS), and/or a mass spectrometer robust enough
for you to transport in (and perhaps even power from) your
car, and as easy to operate as your smartphone? The potential
this would have for metabolomic analysis of samples from huge
numbers of people could drive some incredible work in the
health and medical sciences as well as a range of other
fields. It would be nice to see that in my lifetime. |
Metabolomics Current Contents
|
This
section of MetaboNews is supported by: |
Metabolomics Events
|
| 19-22 Jul 2015 |
ANZSMS25/AOMSC6: The 25th
Conference of the Australian and New Zealand Society
for Mass Spectrometry (ANZSMS) and the 6th
Conference of the Asia and Oceania Society for Mass
Spectrometry Conference (AOMSC) The 2015 conference will include a Metabolomics session sponsored by the Metabolomics Society, featuring Prof Lloyd Sumner. Travel awards are available for Early Career member of the Metabolomics Society and the Australia New Zealand Metabolomics Network (https://sites.google.com/site/anzmn2/travel-awards). For further details, visit http://www.anzsms25.com/. |
| 26-28 Aug 2015 |
Mountain Village Science Series Bio
& Data 2015 The opening lecture will be delivered by Miloš V. Novotný, Director of the Institute for Pheromone Research, Indiana University. Other speakers include Tim Stratton, Reza Salek, Ron Wevers, David Broadhurst and Lennart Eriksson. Abstract submission for both oral and poster presentation is now open, full details of which can be seen here. Please note the closing date for submitting abstracts is Monday 1st June 2015 for oral presentations and Wednesday 1st July 2015 for posters. To register, visit the website at http://www.moviss.org/ |
| 14-17 Sep 2015 |
36th BMSS Annual Meeting 15th-17th
September 2015 Online abstract submission for both oral and poster presentation is now available. Please note the closing date for submitting abstracts is Friday 22nd May 2015. For more information, visit http://www.bmss.org.uk/bmss2015/bmss2015.shtml |
| 21-25 Sep 2015 |
W4M Course 2015: Computational
Metabolomics within the Galaxy Environment on the
Online Workflow4Metabolomics.org (W4M) Resource Program Schedule: http://workflow4metabolomics.org/training/W4Mcourse2015/program Practical Information: http://workflow4metabolomics.org/training/W4Mcourse2015/practical-info Organizing Committee Cécile Canlet, Christophe Caron, Franck Giacomoni, Yann Guitton, Gildas Le Corguillé, and Étienne Thévenot. For further details, visit http://workflow4metabolomics.org/training/W4Mcourse2015. |
| 7-9 Dec 2015 |
MetaboMeeting 2015 Agenda Topics
|
Metabolomics Jobs |
This is a resource for advertising positions in
metabolomics. If you have a job you would like posted in this
newsletter, please email Ian Forsythe (metabolomics.innovation@gmail.com).
Job postings will be carried for a maximum of four issues
(eight weeks) unless the position is filled prior to that
date.
Jobs Offered
| Job Title | Employer | Location | Posted | Closes | Source |
|---|---|---|---|---|---|
| Postdoc |
Max Planck Institute |
Munich, Germany | 22-Jun-2015 |
Metabolomics Society Jobs | |
| Position
as PhD Research Fellow within the field of prostate
cancer metabolism |
Norwegian University of Science and Technology | Trondheim, Norway | 22-Jun-2015 |
26-Jul-2015 |
Metabolomics Society Jobs |
| International Business Development Manager |
BIOCRATES Life Sciences AG | Innsbruck, Austria | 19-Jun-2015 |
19-Jul-2015 | Metabolomics Society Jobs |
| Business Development Manager East Coast |
BIOCRATES Life Sciences AG | Home office based, pref. East Coast | 19-Jun-2015 | 19-Jul-2015 | Metabolomics Society Jobs |
| Director,
Marketing Metabolomics |
Thermo Fisher Scientific | San Jose, California, USA | 18-Jun-2015 |
Until filled |
Metabolomics Society Jobs |
| Three Tenure-Track Faculty Appointments |
Institute of Chemistry, National Autonomous University of Mexico | Coyoacan, Mexico | 12-Jun-2015 |
31-Aug-2015 |
Metabolomics Society Jobs |
| Mass Spectrometry Informatician |
Agios Pharmaceuticals | Cambridge, Massachusetts, USA | 8-Jun-2015 |
Agios
Pharmaceuticals |
|
| Postdoctoral Position, Metabolic Studies
of Cancer Models |
University of California, San Francisco | San Francisco, California, USA | 5-May-2015 |
Until filled |
Metabolomics
Society Jobs |
| Postdoc, Northwest
Metabolomics Research Center |
University of Washington | Seattle, Washington, USA | 17-Apr-2015 |
31-May-2015 or until filled |
Metabolomics
Society Jobs |
| Postdoc, Mitochondria and Metabolism Center | University of Washington | Seattle, Washington, USA | 17-Apr-2015 |
31-May-2015 or until filled |
Metabolomics
Society Jobs |
| Research Assistant I, Metabolomics Core
Facility |
Sanford-Burnham
Medical Research Institute |
Orlando, Florida, USA | 3-Mar-2015 |
Open until filled |
Metabolomics
Society Jobs |
| Postdoctoral Research Fellow, Fernandez
Laboratory, School of Chemistry and Biochemistry |
Georgia Institute of Technology |
Atlanta, Georgia, USA | 19-Feb-2015 |
Open until filled | Georgia Institute of Technology |
|
Ian J. Forsythe Double AB, MSc Editor, MetaboNews Department of
Computing Science
University of Alberta 221 Athabasca Hall Edmonton, AB, T6G 2E8, Canada Email: metabolomics.innovation@gmail.com Website: http://www.metabonews.ca LinkedIn: http://ca.linkedin.com/in/iforsythe Twitter: http://twitter.com/MetaboNews Google+: https://plus.google.com/118323357793551595134 Facebook: http://www.facebook.com/metabonews |
This newsletter is published in
partnership between The Metabolomics Innovation Centre
(TMIC, http://www.metabolomicscentre.ca/) and the Metabolomics
Society (http://www.metabolomicssociety.org)
for the benefit of the worldwide metabolomics
community.
|